Molecular modelers often face a recurring challenge when studying conformational changes in large biomolecular systems: how to generate a realistic trajectory between two known structural states. This issue becomes more complicated when the two states differ significantly, as is the case with the SARS-CoV-2 spike protein transitioning from a closed to an open configuration. In this post, we walk through a practical, reproducible approach to computing such motions using SAMSON, highlighting useful tools and considerations along the way.
We will focus on computing a plausible conformational path between the closed (PDB 6VXX) and open (PDB 6VYB) states of the spike protein. This is particularly relevant in drug discovery and vaccine development where understanding structural flexibility is critical.
Initial hurdles: structure discrepancies
The two conformations originate from different PDB entries and differ in the number of residues, especially in sugar modifications. This discrepancy means a naïve approach to interpolation would produce incorrect results or fail altogether. SAMSON’s workflow accommodates this by allowing editing and harmonization of input structures.
Step-by-step workflow
1. Preprocessing structures:
Using a Python script and SAMSON’s scripting module, bond orders in sugar molecules were corrected in both 6VXX and 6VYB.
Hydrogens were then added, and both structures were energy minimized to prepare them for interpolation. Without this step, path generation could yield energetically unrealistic intermediates.
2. Initial path generation with ARAP:
The As-Rigid-As-Possible (ARAP) Interpolation Path module was used to create a plausible path from the open state to the closed one. ARAP provides rapid first-pass conformational transitions suitable for many large systems. Generation took less than 30 seconds on a laptop.
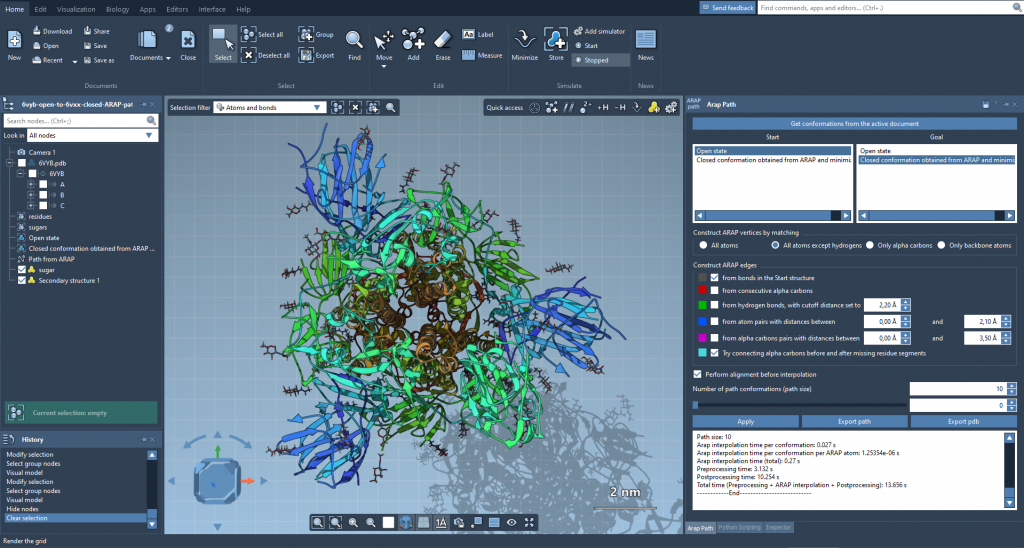
3. Handling residue mismatches:
Due to mismatch in residue numbers, a conformation near the closed state was extracted from the ARAP path, relaxed, and used as the new target structure for a refined ARAP interpolation.
4. Path refinement with P-NEB:
The Parallel Nudged Elastic Band (P-NEB) module was then used to improve the physical realism of the path. P-NEB optimizes intermediate states to lower energy barriers. This step took only about 15 minutes on a regular laptop for the spike protein complex.

Output formats and availability
The computed trajectory is made available in multiple formats: as a set of PDB files, a single PDB file, and in SAMSON’s native format (which includes animation metadata). This allows integration into different modeling workflows.
Most importantly, SAMSON provides these tools free of charge for research on SARS-CoV-2. By following this protocol, you can generate reusable transition paths even for systems with inconsistent formats or structural complexities.
To explore this process in further detail and download all the example files, visit the full documentation page at this link.
SAMSON and all SAMSON Extensions are free for non-commercial use. Download SAMSON at https://www.samson-connect.net.





