Creating libraries of related molecules is a key step in modern drug design and materials science. For molecular modelers, manually generating chemical variants by editing structures can be time-consuming and error-prone. What if you could instantly generate hundreds of structural variants by just defining what to replace, and with what?
This is where the Replace fragments feature of the SMILES Manager Extension in SAMSON comes in. Built on top of the powerful RDKit cheminformatics toolkit, this tool can automate the replacement of molecular fragments, giving you a quick way to generate diverse molecular libraries for screening, visualization, or further computations.
Why fragment replacement?
Subtle changes in molecular structure can have major effects on activity, solubility, and other properties. Medicinal chemists often test varying functional groups, ring systems, or linkers. Instead of modifying molecules one at a time, you can replace defined fragments across multiple molecules in a few steps using SAMSON.
How it works
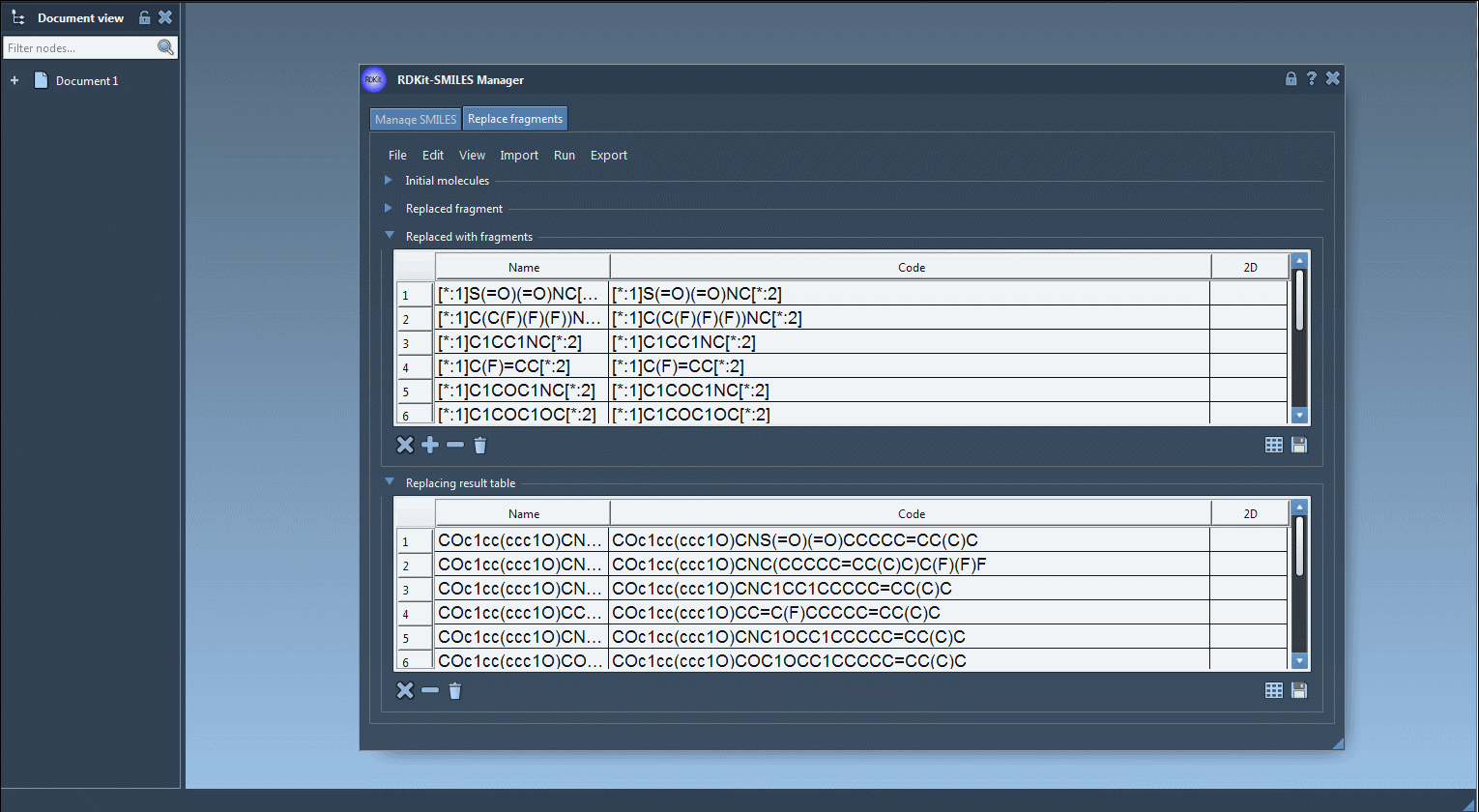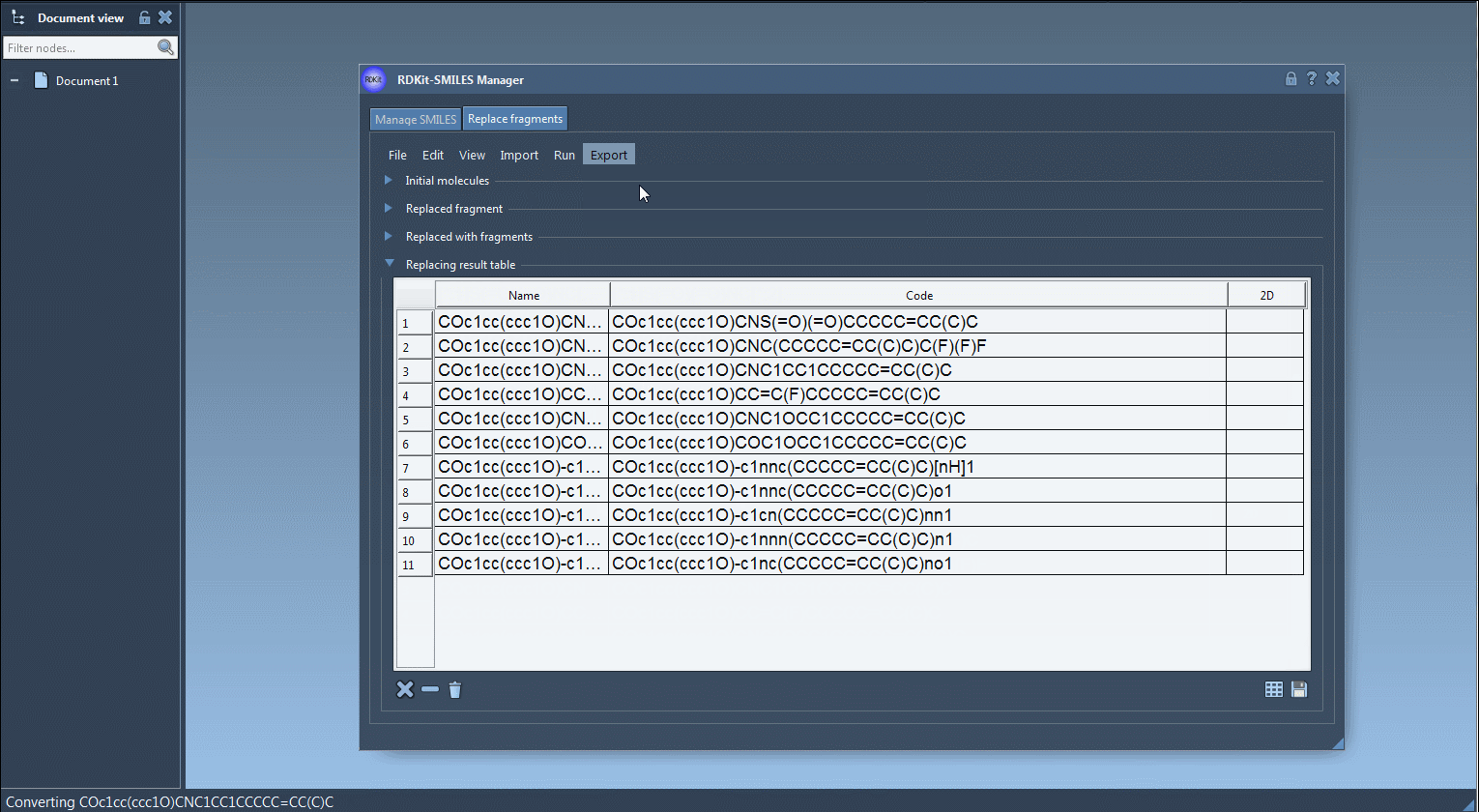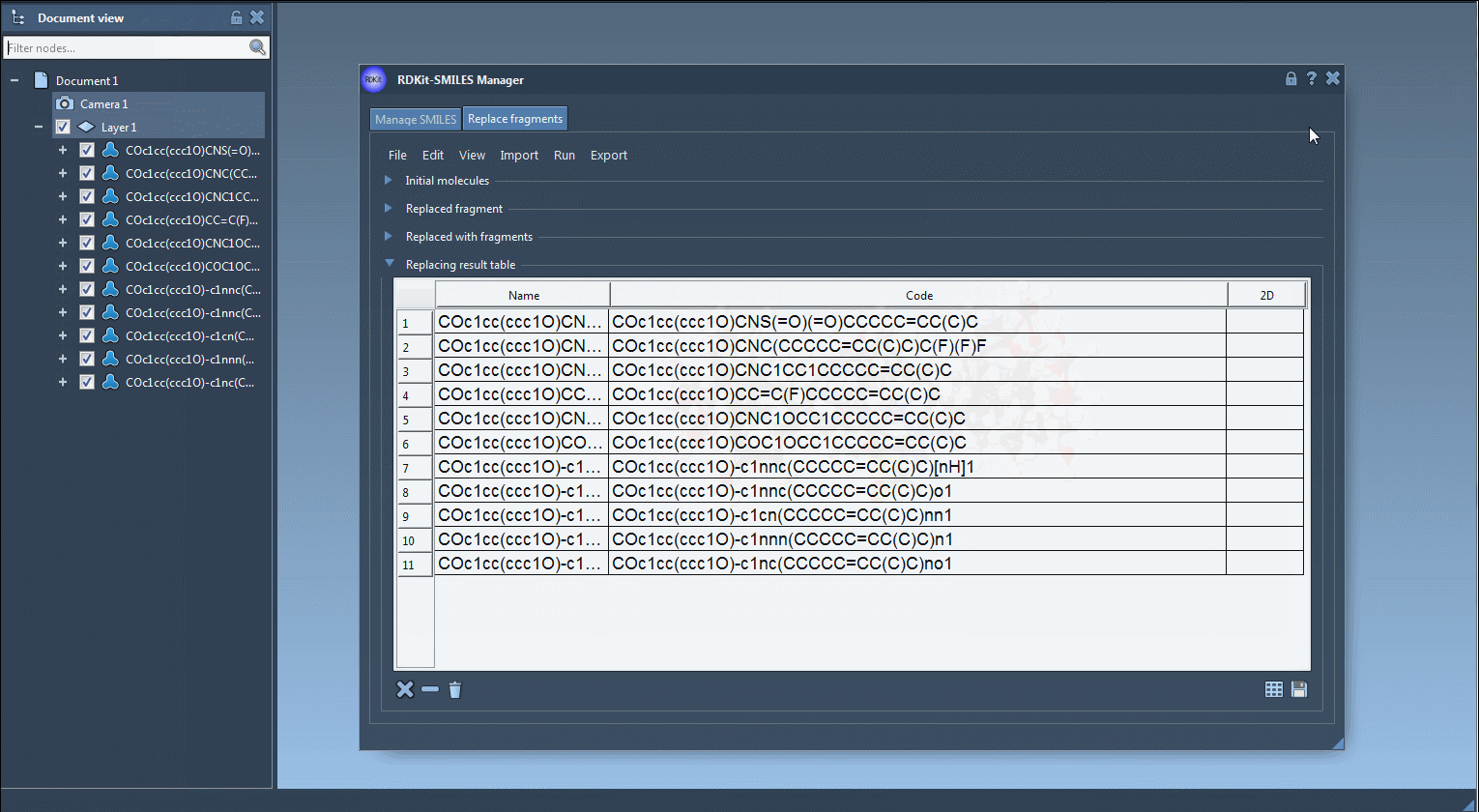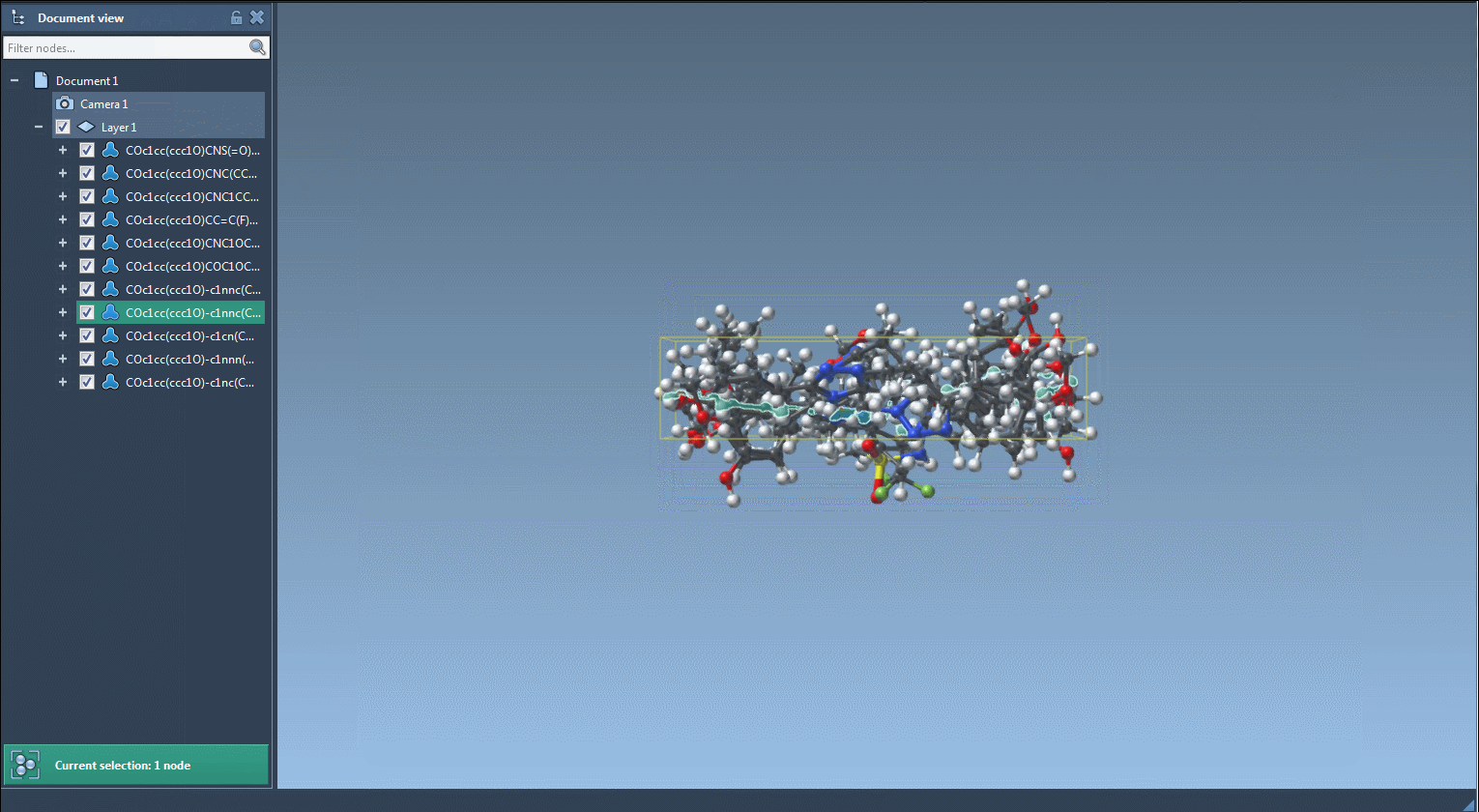
In the Replace fragments tab of the SMILES Manager, the process involves four inputs:
- Initial molecules: The parent compounds where a fragment will be replaced.
- Target fragment: The fragment you want to replace, using SMARTS-like syntax with numbered atoms (e.g.
[*:1]C(=O)NC[*:2]). - Replacement fragments: One or more fragments to substitute in place of the target fragment, using the same connector atom style (e.g.
[*:1]S(=O)(=O)NC[*:2]). - Run: Click to generate the new molecules, which are stored in the Results table and can be viewed or exported.
This approach enables rapid exploration of structural analogs or isosteres, and is especially convenient when analyzing SAR (Structure-Activity Relationships) or synthesizability.
Working example
Let’s walk through a full use case:
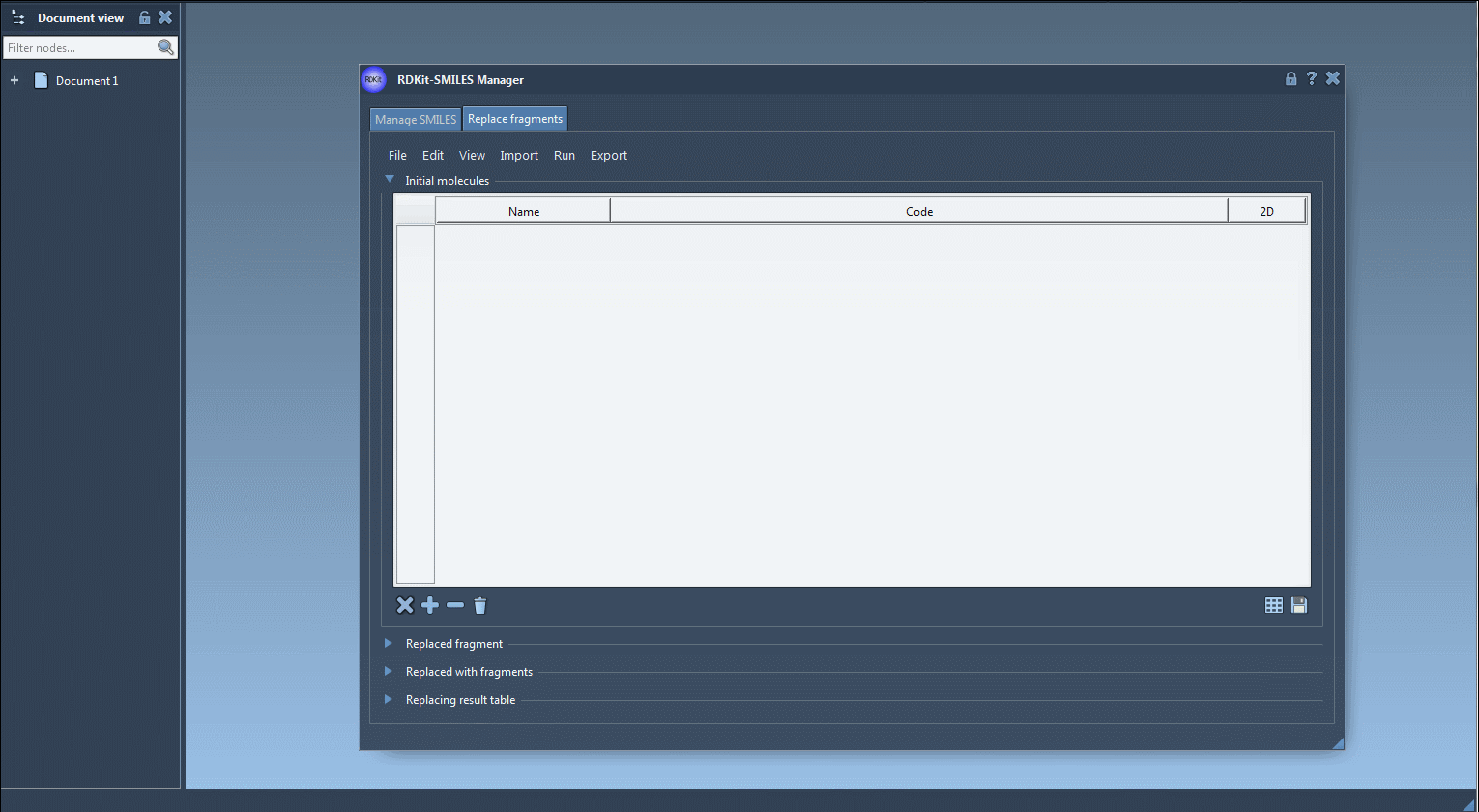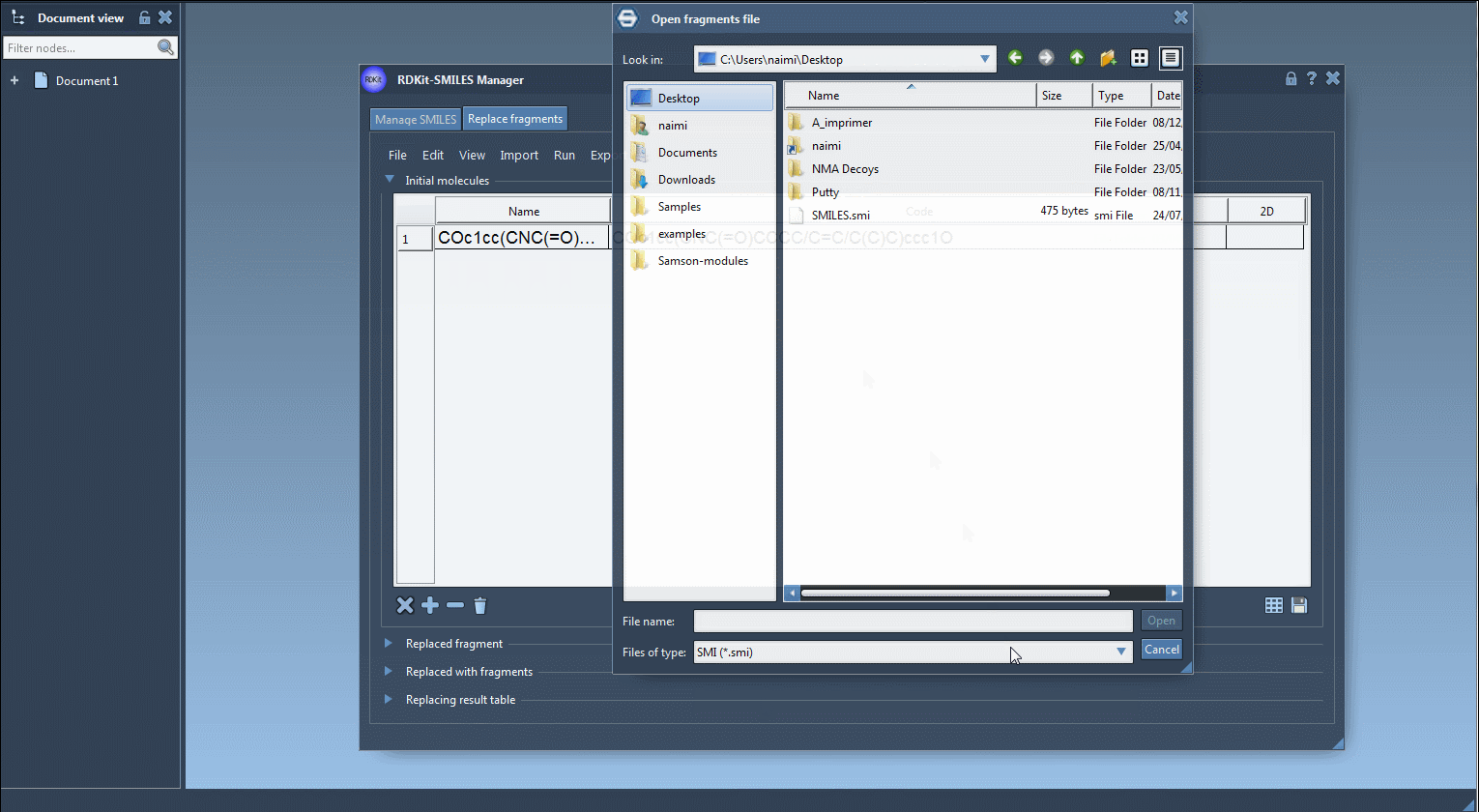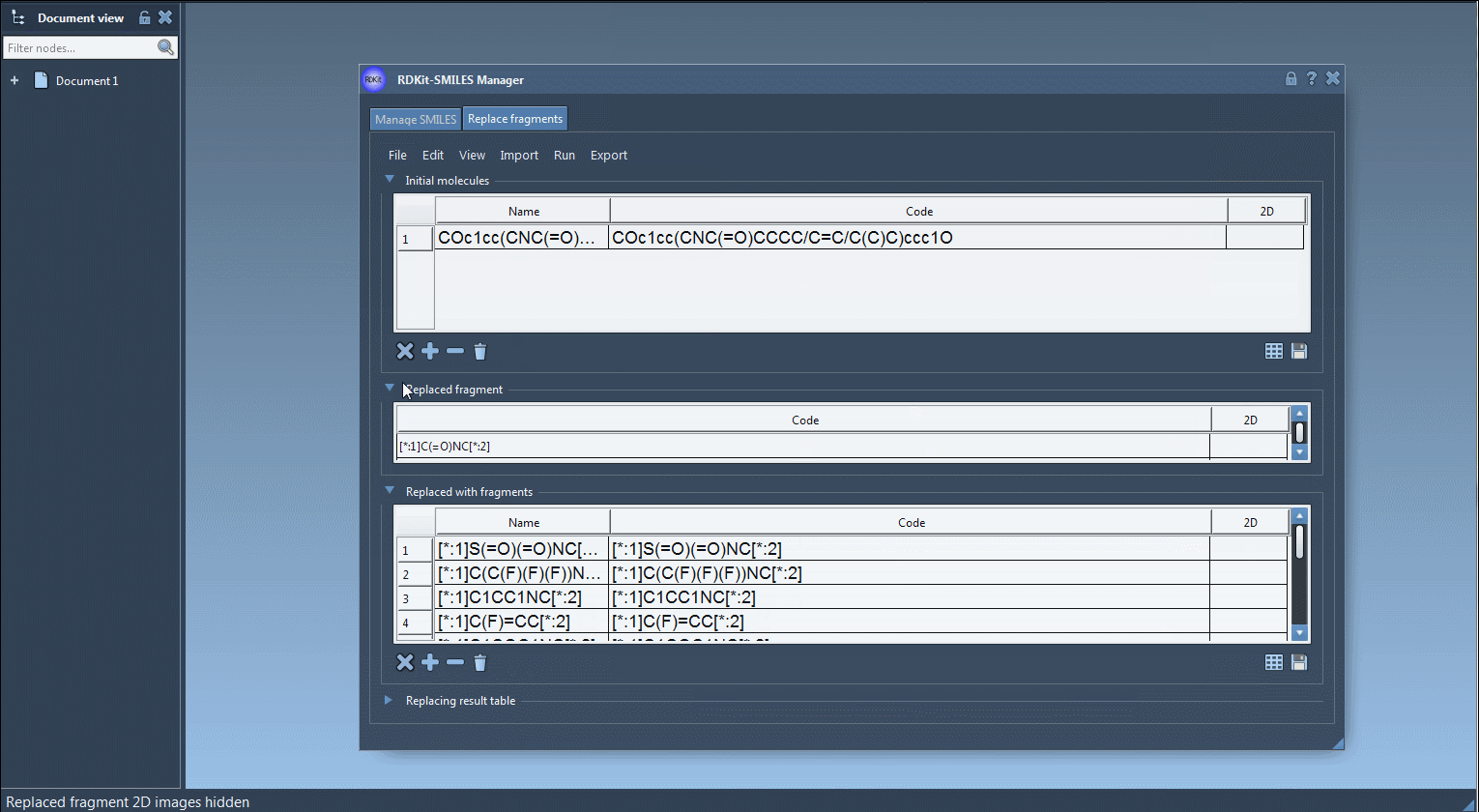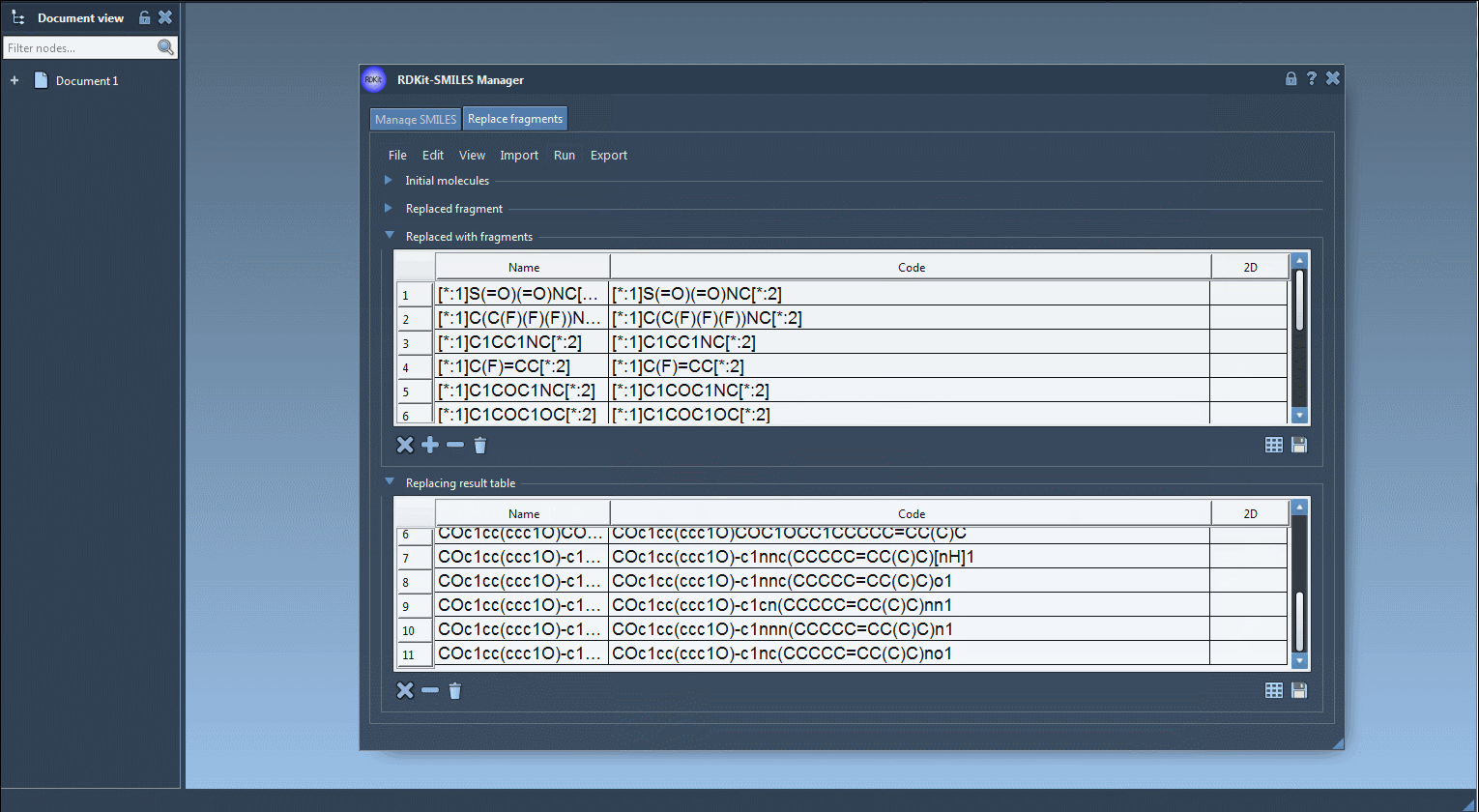
- Input molecule:
COc1cc(CNC(=O)CCCC/C=C/C(C)C)ccc1O - Fragment to replace:
[*:1]C(=O)NC[*:2] - Replacement fragments (a few examples):
[*:1]C(C(F)(F)(F))NC[*:2][*:1]C1CC1NC[*:2][*:1]C1COC1OC[*:2]
Once you click Replace fragments, the Results table will populate with all new combinations. You can instantly generate 3D structures, save them as images, or explore them in SAMSON’s molecular viewer.
Tips for effective use
- Use wildcard atoms like
[*:1]and[*:2]for flexible connection points. - Save the result as a grid image to visually compare your library, with names shown or hidden.
- Preview 2D depictions before generating 3D models to filter desired variants.
This powerful feature saves time and reduces manual editing, making it easier to explore chemical space without switching tools or writing code.
To learn more and see additional examples, visit the full tutorial on fragment replacement at SAMSON’s documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at www.samson-connect.net.





