When working with large biological assemblies—whether viral capsids, multimeric proteins, or supramolecular complexes—understanding the underlying symmetry is more than a theoretical curiosity. It can unlock practical efficiencies in molecular modeling, validation, and simulation setup. But what happens when a structure appears to have multiple symmetrical arrangements? How do you choose which group is most appropriate for your modeling task?
The Symmetry Detection extension in SAMSON offers automatic identification of cyclic, dihedral, and cubic symmetries in biological assemblies. For small complexes, a single dominant symmetry is often detected. However, for large structures, it’s common to see more than one candidate symmetry group returned. This blog post explains how to interpret these cases and choose—manually or automatically—the most suitable group for your project.
Why Multiple Symmetry Groups May Be Returned
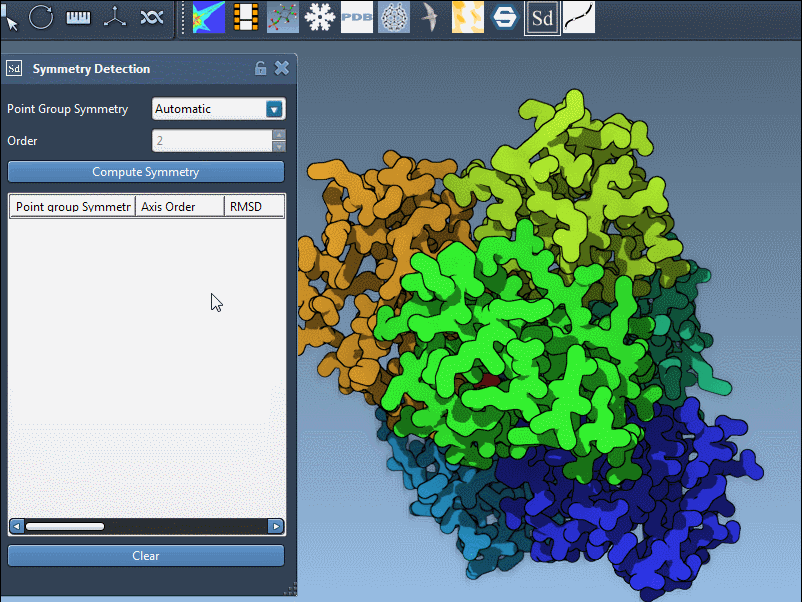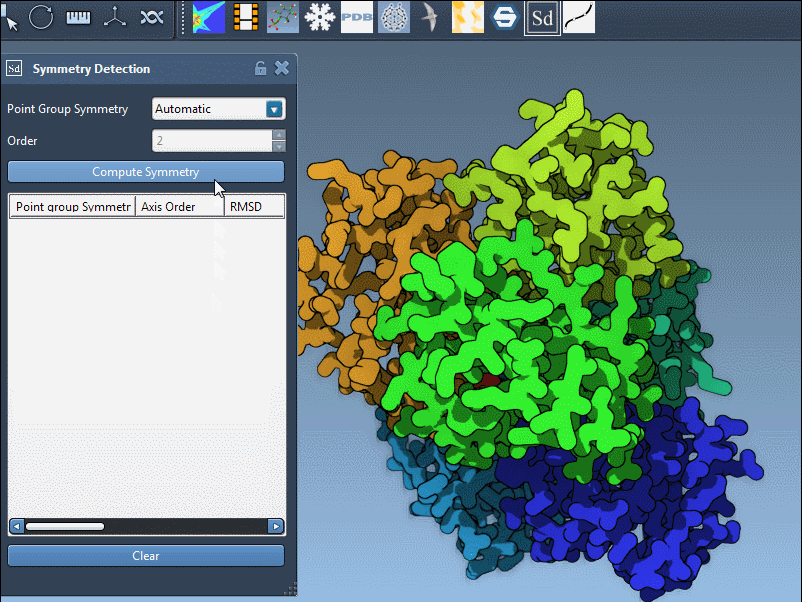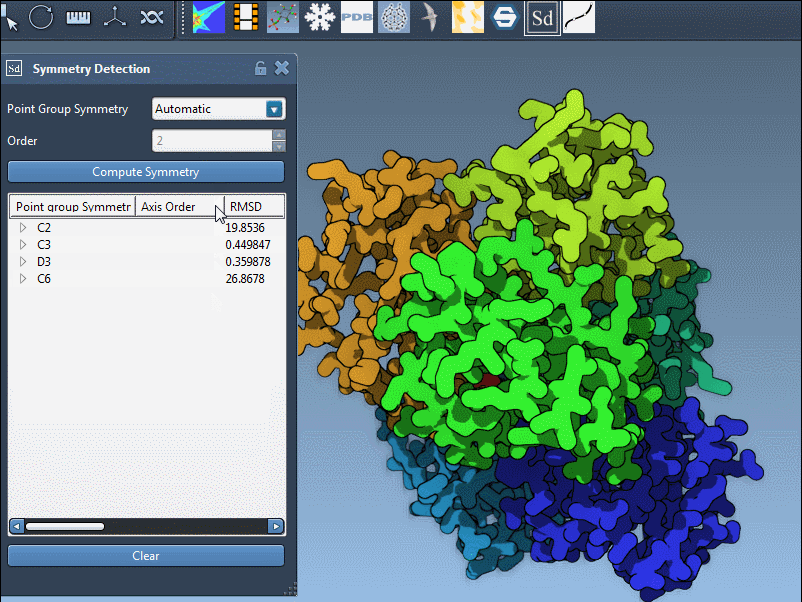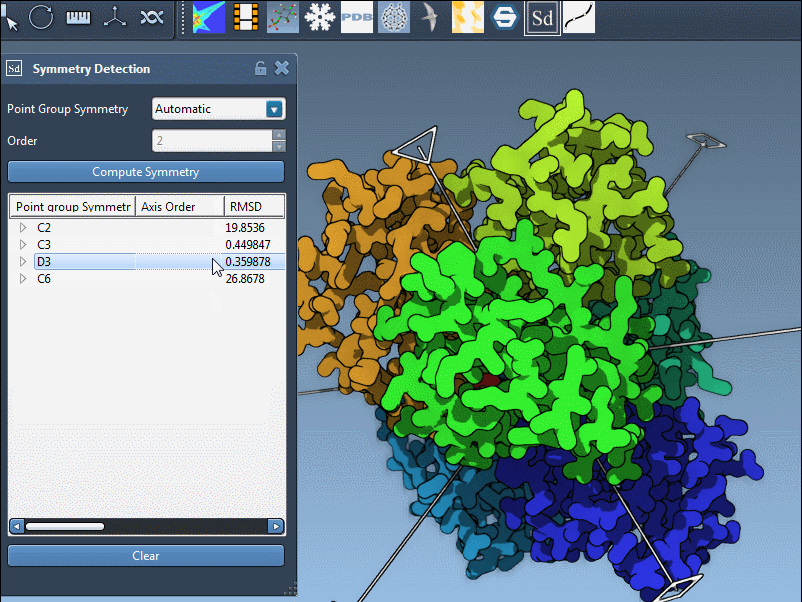
Biological assemblies, especially large ones, may contain substructures with different local symmetries or may approximate multiple global symmetries. When SAMSON analyzes such a structure, it evaluates all plausible arrangements and returns the groups that provide a good fit to the molecular geometry. The result is a list of symmetrical arrangements, each scored using a root square mean deviation (RMSD).
Choosing the Best Symmetry Group
Here are two simple but effective criteria you can use:
- Prefer higher-order groups that explain more of the molecule with fewer unique units.
- Lower RMSD values suggest a better geometric match, implying a more accurate symmetry representation.
In SAMSON, you can visually inspect the different symmetrical options:
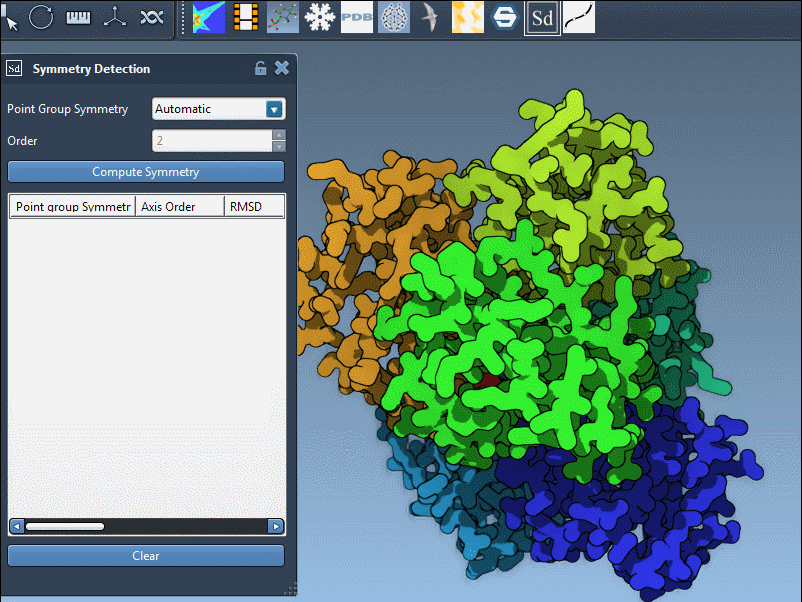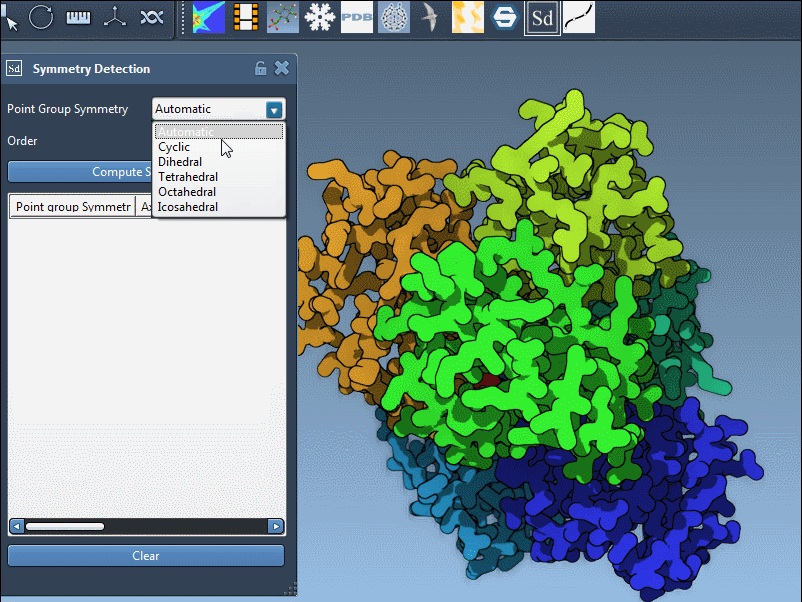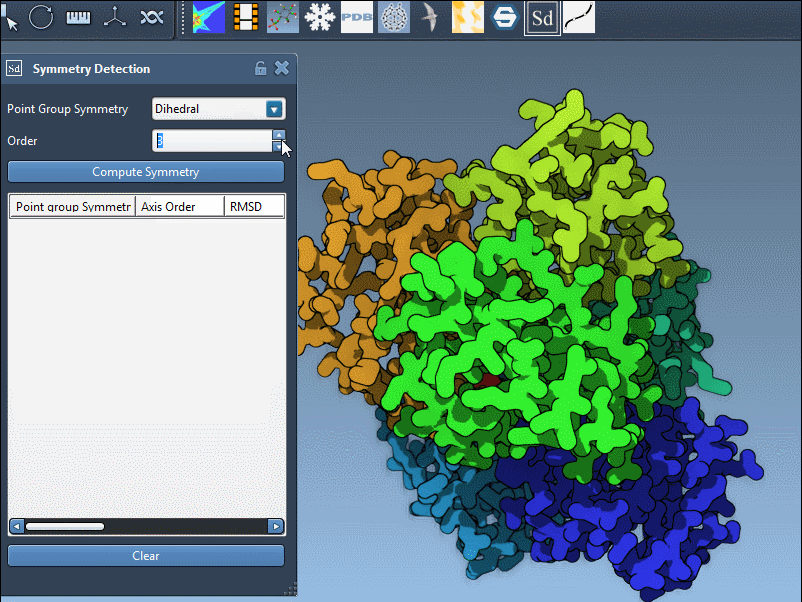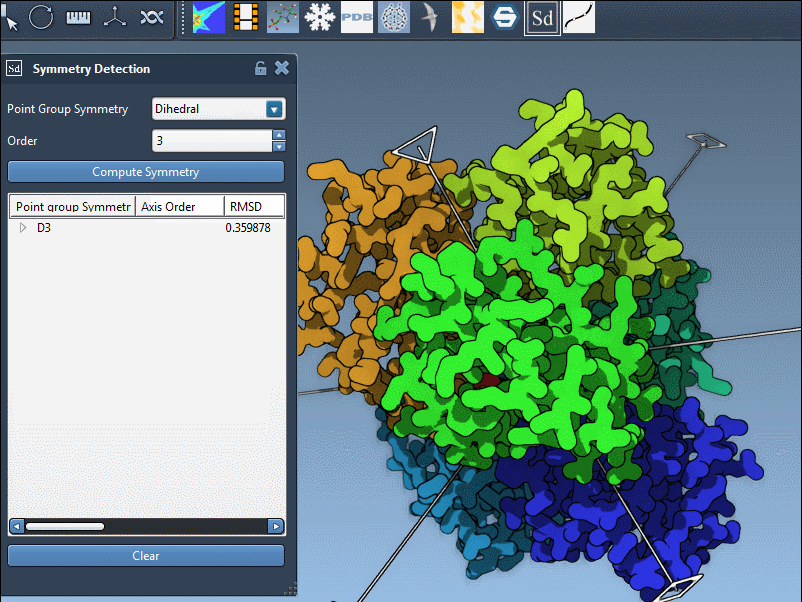
- Click a group to highlight its main axis in the viewport.
- Hover over or expand the group to see individual axes and RMSD values.
- Use single-click to highlight an axis or double-click to align the camera with it.
Here’s an example from the PDB structure 1B4B. The extension suggests a D3 (dihedral order 3) symmetry. Selecting this group shows a clear, repeatable threefold axis that matches the biological arrangement well:
Specifying a Group Manually
If your prior knowledge points you toward a specific group (e.g., based on literature or experimental design), you can manually assign it using the dropdown menus in the extension panel. This is particularly useful for validating your results or trying alternate hypotheses. In the example below, we select the D3 group directly:
Tips for Better Judgment
Comparing options visually and numerically helps you decide which symmetry is most useful for your goals. Not sure? Try answering these small questions:
- Which group yields the lowest RMSD?
- Does the highlighted axis align well with known structural features or binding interfaces?
- Will this group reduce computational cost by allowing simulation of just an asymmetric unit?
Conclusion
Dealing with multiple possible symmetry groups can seem complex, but SAMSON gives you both the automation and control to make informed decisions. Whether you’re modeling mutagenesis, simplifying coarse-grained simulations, or validating structural data, informed symmetry selection can save time and increase modeling fidelity.
To learn more, visit the full documentation page: Symmetry Detection in Biological Assemblies.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





