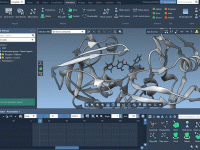
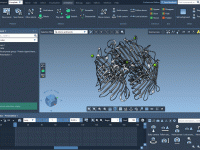
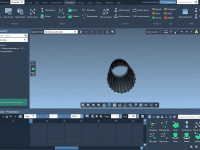
Simplify Molecular Modeling with Undo and Redo in SAMSON
Create Seamless Molecular Animations with the Hold Camera Feature
Mastering the Zoom Camera Animation in Molecular Modeling
Understanding and Using Residue Attributes in Molecular Design
Molecular modelers often face the challenge of efficiently analyzing and manipulating specific residues within complex molecular structures. SAMSON, the integrative molecular design platform, provides a powerful solution. With its Node Specification Language (NSL), SAMSON offers a structured system to define…
A Clear Roadmap to Preparing Coarse-Grained Systems in GROMACS Wizard
For molecular modelers tackling complex simulations, preparing coarse-grained (CG) systems can feel like navigating a maze. Whether you’re handling large biomolecules or experimenting with customized solvent models, the need for clear, efficient preparation workflows is critical. Fortunately, the GROMACS Wizard…