When setting up molecular dynamics simulations of membrane proteins, one of the most repetitive and error-prone steps is placing a lipid layer or bilayer around the protein. Aligning the protein, choosing the right lipid orientation, adjusting the box dimensions, and ensuring that no molecules overlap— molecular modelers often spend hours manually tweaking these parameters.
Fortunately, SAMSON’s Molecular Box Builder extension simplifies this process considerably. Here’s a walkthrough for generating a lipid layer around a membrane protein, automatically ensuring correct placement and orientation.
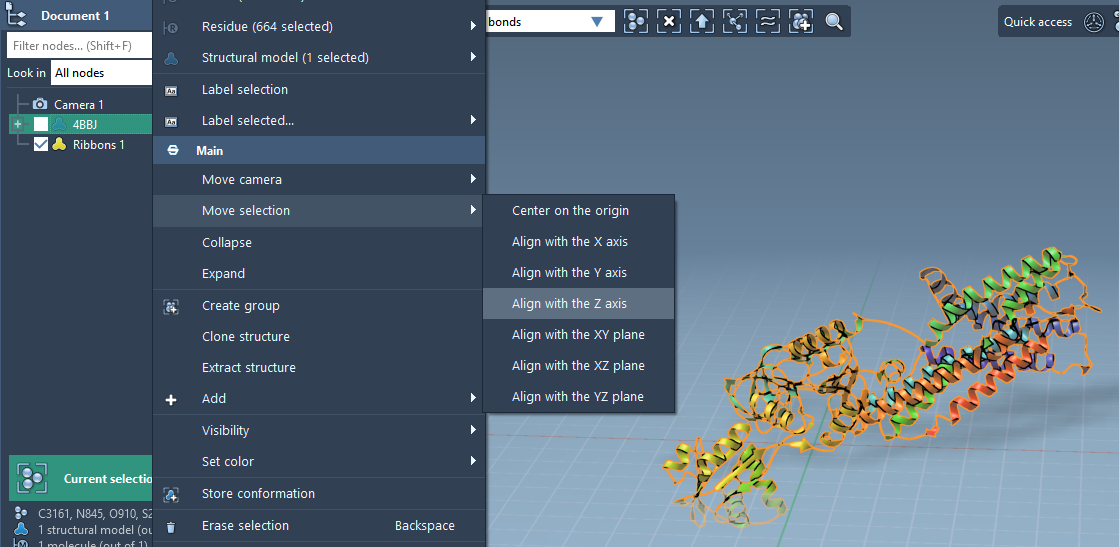
Reorient the Protein for Membrane Placement
Begin by importing and selecting your protein in the Document view. To align it perpendicularly to the membrane plane:
- Right-click the protein.
- Choose Move selection > Align with Z axis.
- Then select Move selection > Center on the origin.
This positions the protein along the Z-axis, a standard orientation for membrane embeddings and simulation prep.
Define the Lipid Molecule to Insert
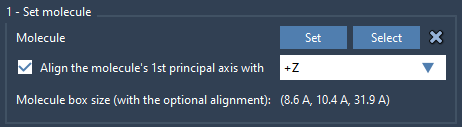
Next, import your lipid molecule. Once added:
- Select the molecule and click Set in the Molecular Box Builder app.
- Choose to align the molecule with the
+Zaxis, so the lipid tails and head groups are oriented correctly.
Define the Packing Box Around the Protein
Open the Set Box section of the app. This is where most of the trial-and-error is usually spent, but here it’s simplified:
- Center the box on the aligned protein.
- Adjust the box dimensions to contain a single molecular layer (typically, a narrow Z-axis and larger X and Y values).
- If needed, specify a margin between inserted molecules to adjust density.
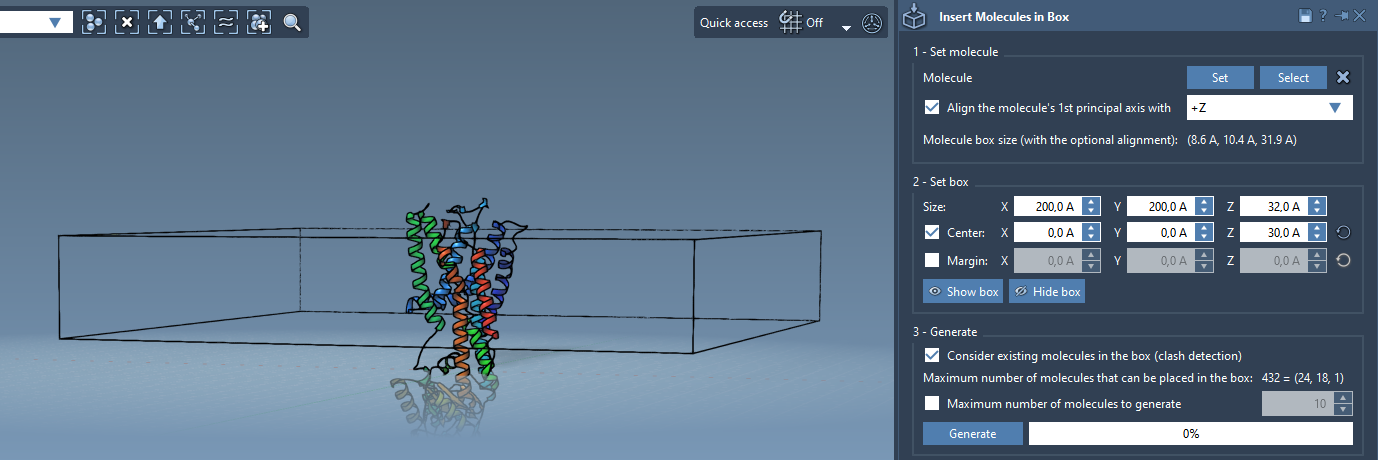
Generate the Lipid Layer
This is as simple as clicking Generate — but make sure to enable Consider existing molecules in the box. This option ensures that the lipid molecules are inserted only in available space and don’t clash with the protein.
The result is a single layer of lipids, oriented correctly and packed around the protein structure.

Bonus: Build a Bilayer
Creating a bilayer is just a few additional steps:
- Generate the first layer with
+Zalignment. - Shift the box center downward in Z.
- Add the second layer with
-Zalignment.
Now the protein sits in a membrane bilayer — no manual editing required.
This workflow removes manual steps and eliminates common alignment mistakes, making it easier to start simulations with correct initial conditions.
To learn more about the Molecular Box Builder, explore the full documentation at this tutorial page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





