Building lipid layers around membrane proteins is a routine yet tedious task in molecular modeling. Ensuring correct orientation and placement, preventing atomic clashes, and avoiding manual repetition are often time-consuming steps. Fortunately, SAMSON’s Molecular Box Builder provides an efficient way to create lipid monolayers and bilayers around proteins—using a few intuitive steps that spare you the guesswork.
In this post, we’ll walk through how to generate a single or double lipid layer surrounding a protein structure. This can save hours of manual setup and is a helpful preparation step before running simulations on membrane proteins or receptor-lipid interactions.
Step 1: Prepare and Align Your Protein
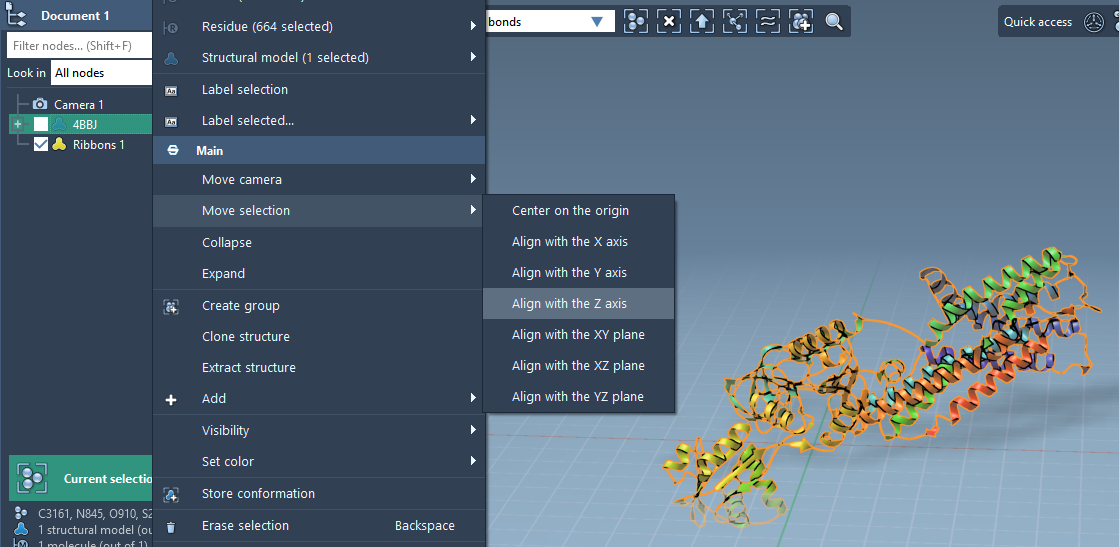
Start by importing your protein of interest into SAMSON—here, for example, the copper-transporting PIB-ATPase (PDB ID: 4BBJ). Proper protein orientation is essential for meaningful bilayer placement. To align your protein along the Z-axis, simply:
- Right-click the protein in the Document view.
- Select Move selection > Align with Z axis.
- Then Move selection > Center on the origin.
Step 2: Add and Align the Lipid Molecule
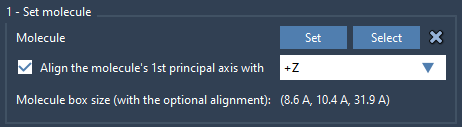
Next, import a lipid molecule (e.g., POPC), which will serve as your layer component. Once it is loaded:
- Select the lipid and click Set in the Molecular Box Builder interface.
- Align its principal axis with the
+Zaxis using the orientation setting.
Step 3: Define the Box
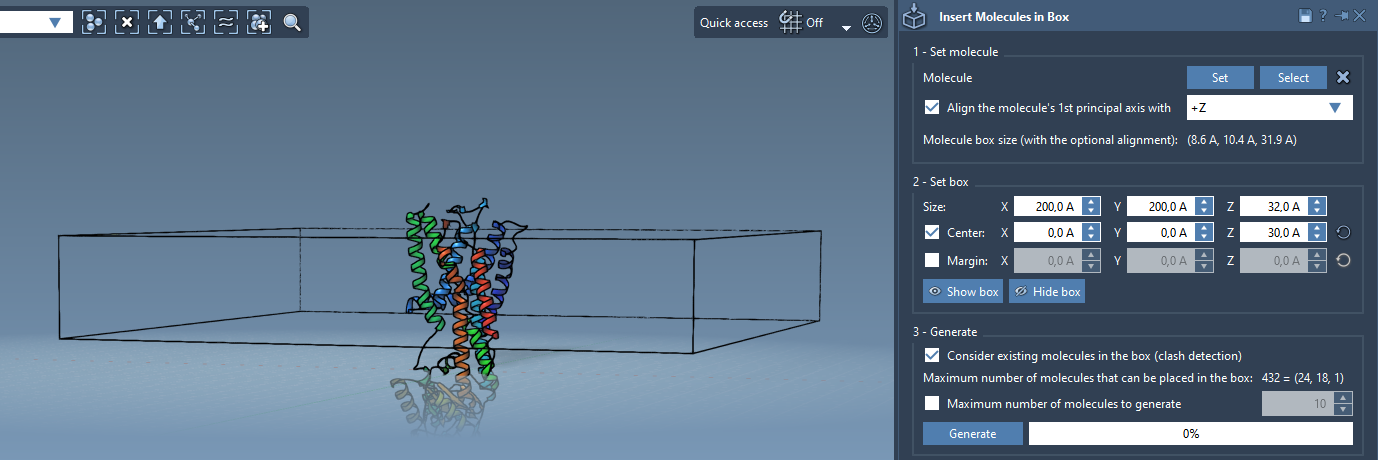
The next step is defining the volume in which the lipid copies will be placed. To do this:
- Center the box around the protein.
- Adjust its size to span the XY dimensions of the desired membrane and Z-thickness of a monolayer.
- If needed, add or reduce the margin between molecules to alter density.
Step 4: Generate the Lipid Layer
Now enable the option Consider existing molecules in the box to prevent overlap with the protein. Then click Generate. The lipid molecules will automatically populate the available regions around the protein.

Optional: Generate a Lipid Bilayer
To construct a full bilayer:
- First, create the top leaflet by adding the
+Z-aligned lipid layer. - Then shift the box center below the protein in Z.
- Switch the lipid alignment to
-Zand generate the bottom leaflet.
This two-step process gives you a complete bilayer surrounding your protein in a structurally consistent way—with minimal manual effort.
This approach eliminates time-consuming and repetitive copy-paste tasks for assembling membranes by hand. Combined with further extensions such as GROMACS Wizard for equilibration and dynamics, this workflow is practical and accessible for both entry-level and experienced modelers.
To learn more, refer to the original Molecular Box Builder tutorial: https://documentation.samson-connect.net/tutorials/molecular-box-builder/molecular-box-builder/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





