Membrane proteins are notoriously tricky to simulate, in part because preparing a realistic environment around them can be a frustrating and time-consuming process. Modeling a lipid bilayer around a transmembrane protein usually requires specialized tools, scripting skills, and plenty of patience to resolve overlaps and positioning issues.
With the Molecular Box Builder extension in SAMSON, you can now generate lipid layers around membrane proteins visually, in minutes, and without scripting. This blog post walks you through creating a lipid monolayer—and even a full bilayer—around a membrane protein structure like 4BBJ, a copper-transporting PIB-ATPase.
Align the Protein
Before surrounding a protein with lipids, it’s important to orient it consistently. Typically, researchers align the transmembrane axis with the Z-axis, enabling bilayer placement orthogonal to this direction.
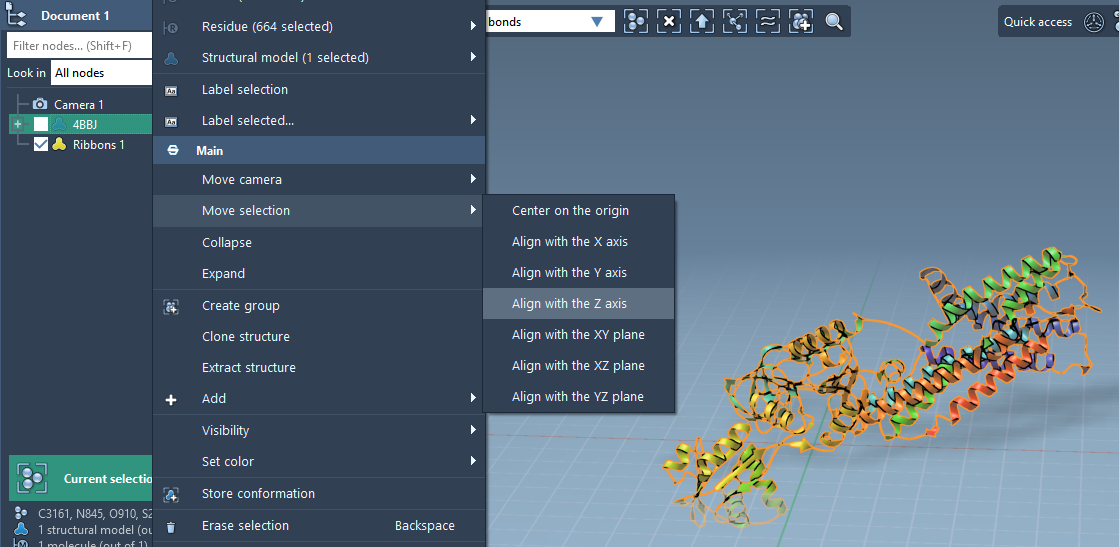
- Right-click on the protein in SAMSON’s Document view.
- Select Move selection > Align with Z axis.
- Then choose Move selection > Center on the origin.
Set the Lipid Molecule
Choose a lipid to build your layer. This could be POPC, DPPC, or any custom lipid you’ve modeled.
- Import the lipid molecule into your SAMSON document.
- Select it and open the Molecular Box Builder.
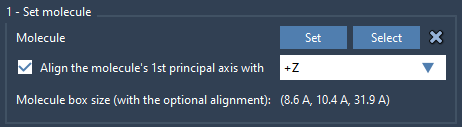
- Click Set to define it as the molecule to populate the box.
- Align the molecule’s main axis to the
+Zdirection for correct bilayer orientation.
Define the Box Around the Protein
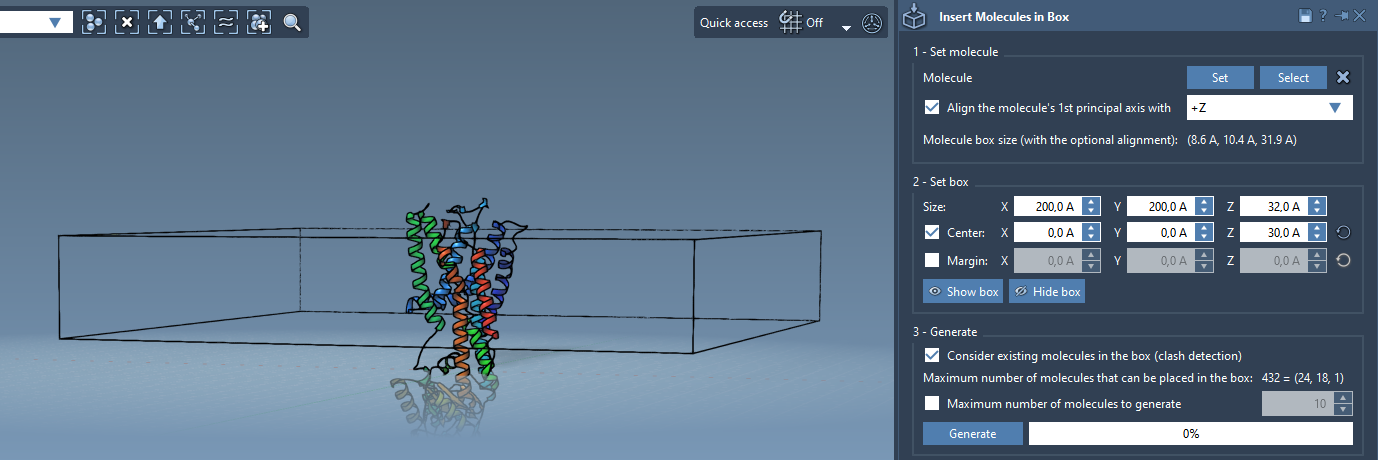
The lipid molecules are arranged within a customizable 3D box. For a realistic monolayer, size the box to match the protein’s horizontal span and the desired thickness along the Z-axis.
- Center the box around the protein model.
- Adjust the box dimensions in
XandYdirections to cover the membrane plane. Use a small Z-height (e.g., ~5 Å) for a monolayer. - Set a suitable margin between lipids to prevent atomic overlaps. A margin between 1.5 and 3 Å usually works well.
Generate a Lipid Layer
Once your box is defined, simply populate it with your lipid molecule:
- Enable the checkbox Consider existing molecules in the box to avoid placing lipids where the protein already occupies space.
- Click Generate.
The result is a lipid monolayer distributed around your protein model—no scripting required.

Optional: Creating a Bilayer
Want both extracellular and intracellular sides covered?
- After adding the first lipid layer aligned to
+Z, shift the box slightly downward along the Z-axis. - Change your lipid alignment to
-Zand click Generate again.
Now both sides of the protein are covered with lipids:

Finishing Steps
After generating the lipid environment, you might want to:
- Use the PDBFixer extension to add solvent and ions.
- Minimize and equilibrate using the GROMACS Wizard.
This visual, flexible approach to lipid layer modeling can significantly reduce setup time and learning curves for early-stage simulations of membrane systems. To learn more, visit the full Molecular Box Builder documentation.
Note: SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at www.samson-connect.net.





