When working with proteins in PDB format, one of the recurring challenges is understanding how a structure fits into its larger biological context. Many deposition files contain only an asymmetric unit—just a fragment of the full biological assembly. This often leads to confusion: is this monomer the complete structure, or is it just one part of a larger complex?
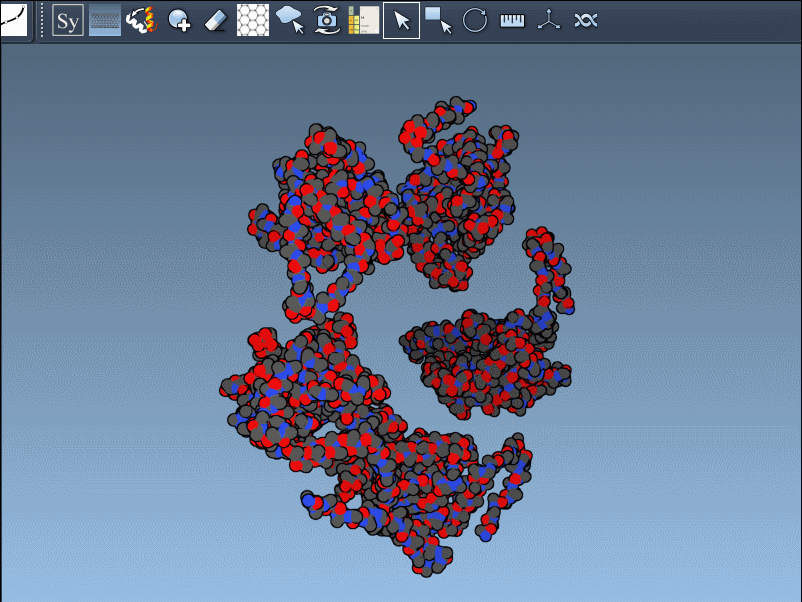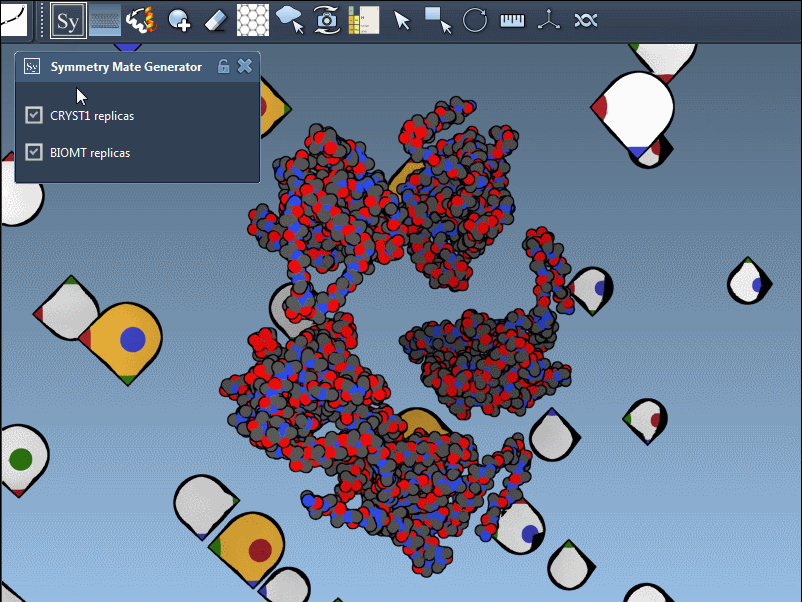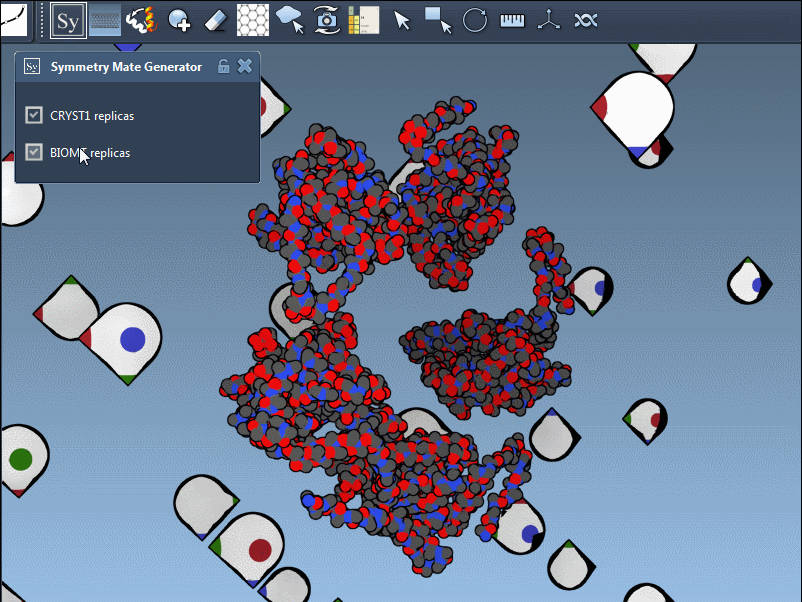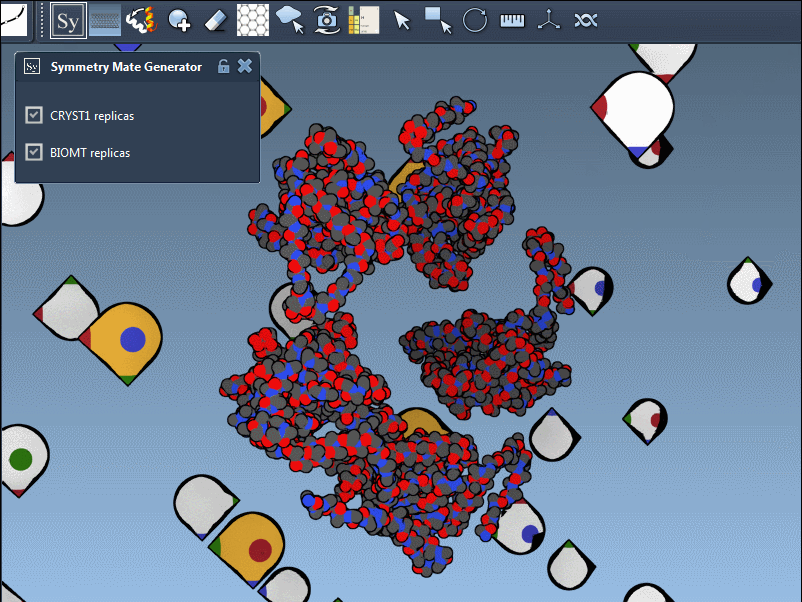
The Symmetry Mate Editor in SAMSON is designed to bridge this gap. It lets you interactively generate and visualize symmetry mates—replicas of a structure inferred from crystallographic (CRYST1) or biological assembly (BIOMT) annotations. What makes this feature especially helpful is how intuitive and visual it is: you can explore potential assemblies simply by hovering, scrolling, and clicking.
What Are Symmetry Mates, Exactly?
In many PDB files, detailed annotations describe the transformations needed to reconstruct the full biological assembly. These transformations fall into two categories:
- CRYST1 records describe crystal packing, often used to visualize repetitive units in the lattice.
- BIOMT records refer to biologically relevant assemblies—actual oligomeric forms observed in vitro or in vivo.
Understanding both symmetry representations is key to protein modeling, particularly for applications like interface analysis or nanostructure design. The Symmetry Mate Editor makes this process both accessible and reversible.
How It Works in Practice
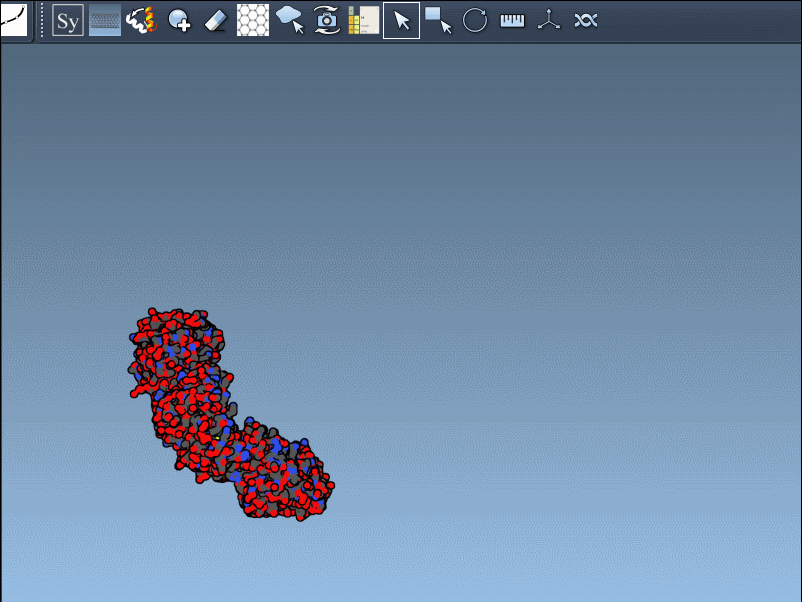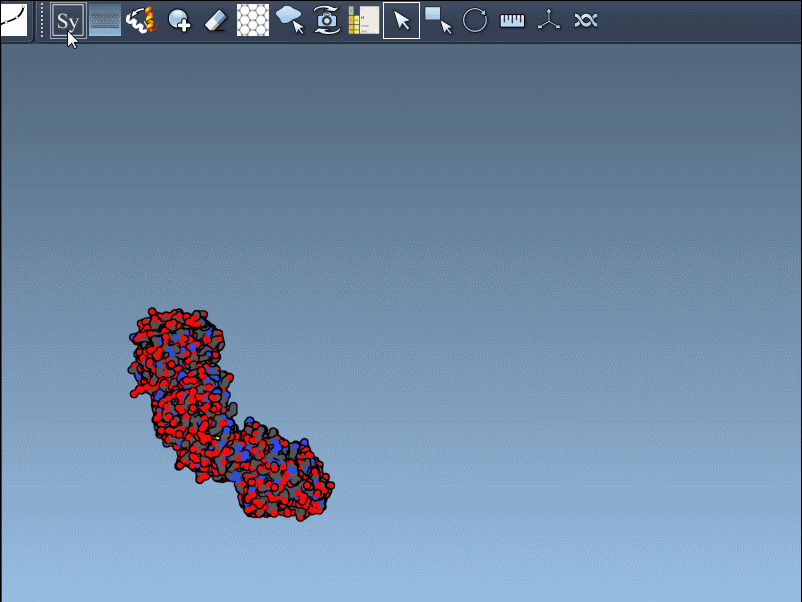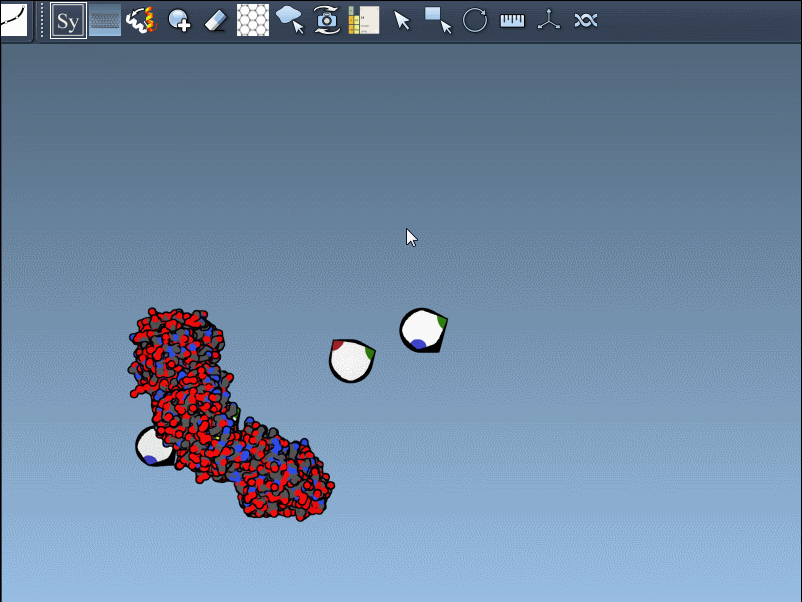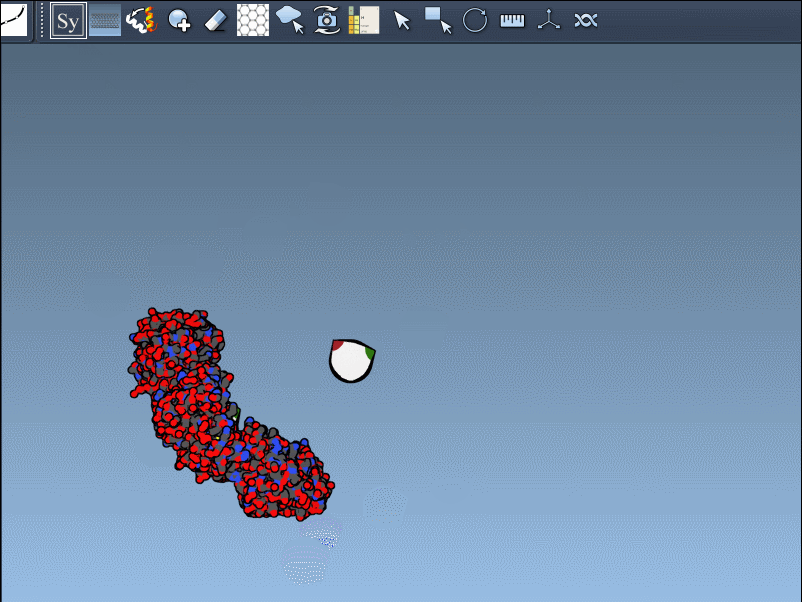
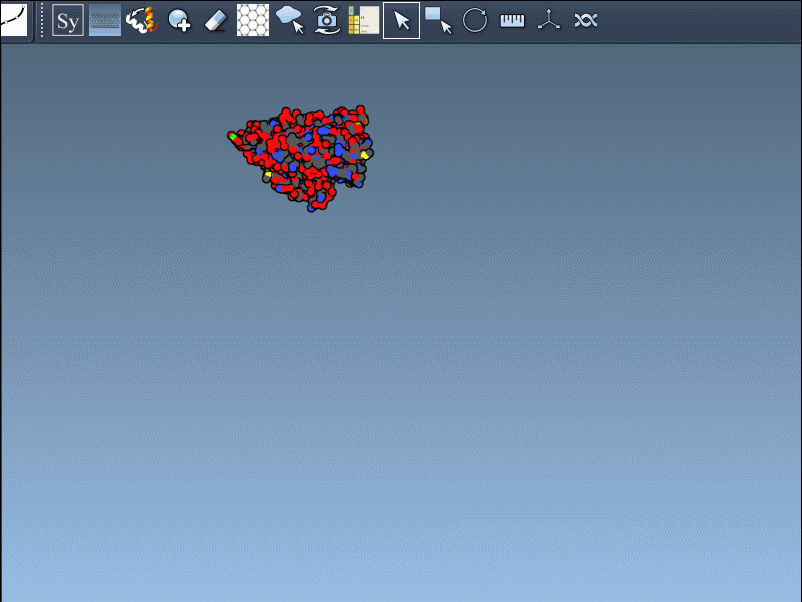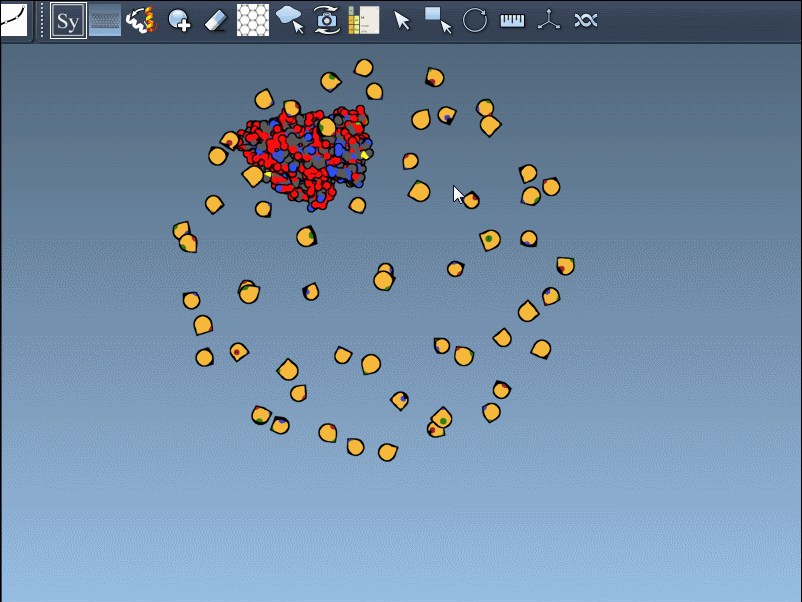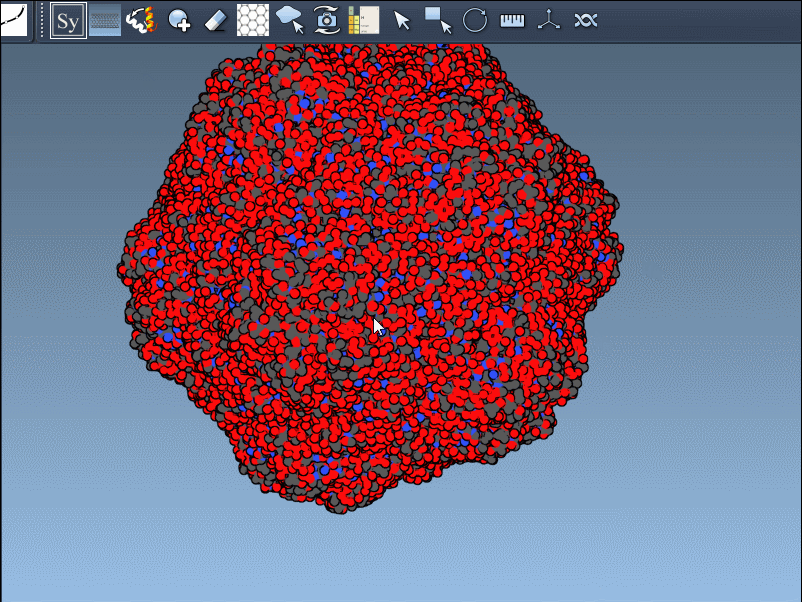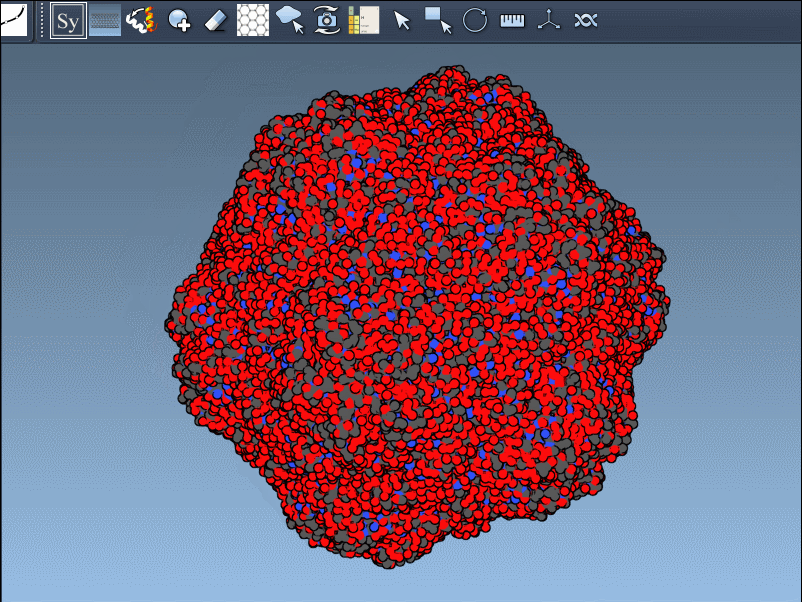
Once you activate the Symmetry Mate Editor from SAMSON’s interface (via the Find Everything shortcut or the editor menu), you’ll see control nodes appear in the 3D viewport. Each node corresponds to a possible symmetry transformation for the currently loaded protein structure.
- Hover over a node to preview a mirror or rotated version of the original structure.
- Left-click to generate a permanent replica in the workspace.
- Use Ctrl/Cmd + scroll to scale the number of visible symmetry nodes—more nodes will appear or disappear accordingly.
- Undo (Ctrl/Cmd + Z) will remove modifications or unwanted replicas.
The interactive visualization is particularly helpful for large assemblies. You can even preview all possible symmetry mates at once—press and hold Ctrl/Cmd before hovering and clicking a node. This is useful when trying to reconstruct full viral capsids, symmetric protein scaffolds, or just exploring potential interfaces for docking studies.
Why It Matters
This approach solves a practical problem: quickly determining how your protein of interest participates in larger assemblies, without needing to manually process transformation matrices or load external software. For structural modelers, it means faster iteration. For educators, it’s a hands-on way to show how molecules fit in 3D space. For designers, it enables rapid testing of ideas involving symmetry or modular protein design.
You can also visually distinguish between CRYST1 and BIOMT entries. In the Symmetry Mate Editor, white nodes correspond to crystallographic symmetry, and yellow nodes to biological assembly symmetry. This helps clarify whether you’re dealing with biologically meaningful interfaces or simply packing contacts.
To learn more about symmetry mate generation in SAMSON, visit the full documentation page: https://documentation.samson-connect.net/tutorials/symmetry/generating-symmetry-mates/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





