When working with protein structures from the Protein Data Bank (PDB), modeling biological assemblies usually requires rebuilding symmetry-related units based on transformation matrices encoded in the structure file. This can often be a tedious task — and yet it’s essential: understanding the assembly of a protein in its crystal or biological context is crucial for studying its function, interactions, and dynamics.
The Symmetry Mate Editor in SAMSON offers a visual and interactive way to explore and generate these symmetry mates. If you’ve been mapping out binding sites, simulating protein complexes, or designing symmetric biomolecular structures, this feature might save you time and effort — and add insight to your workflow.
How It Works: A Smooth Preview-and-Click Experience
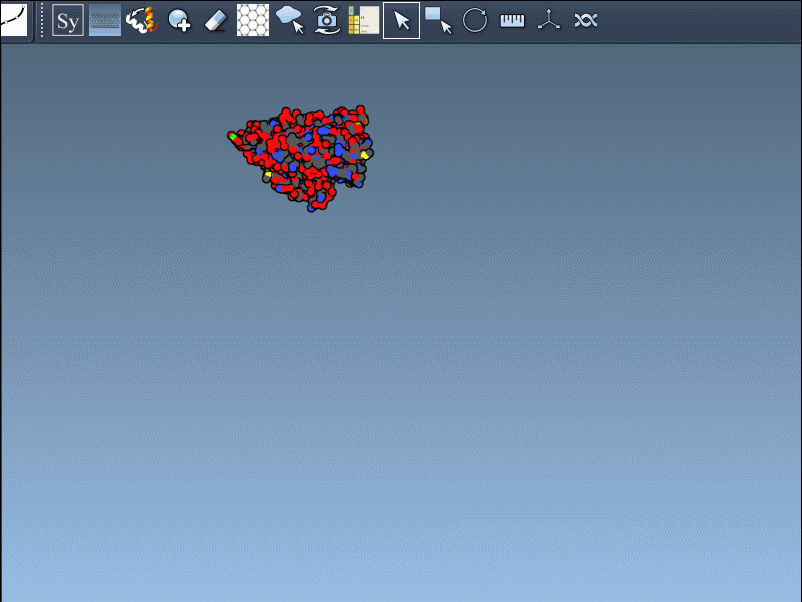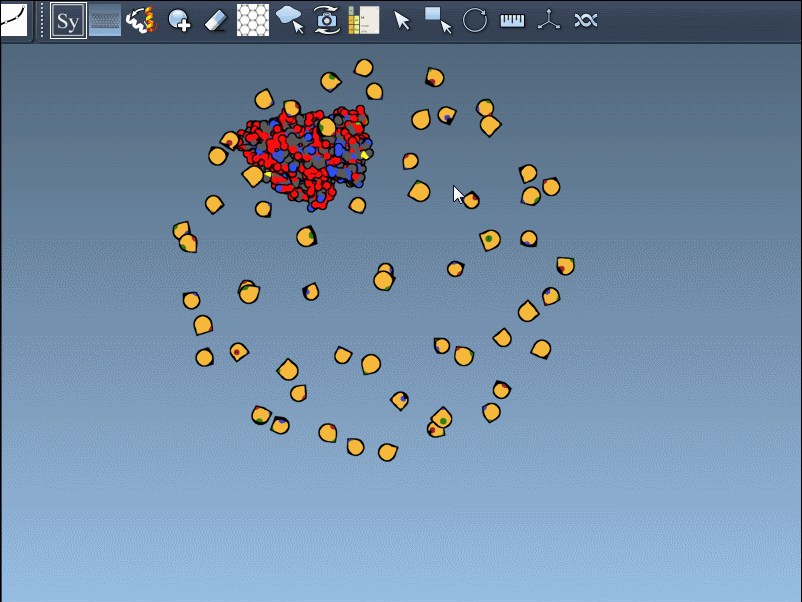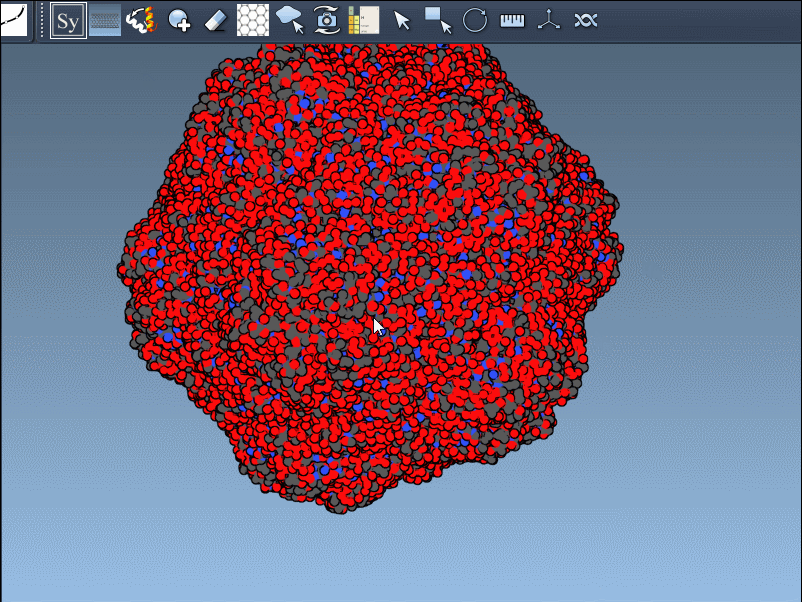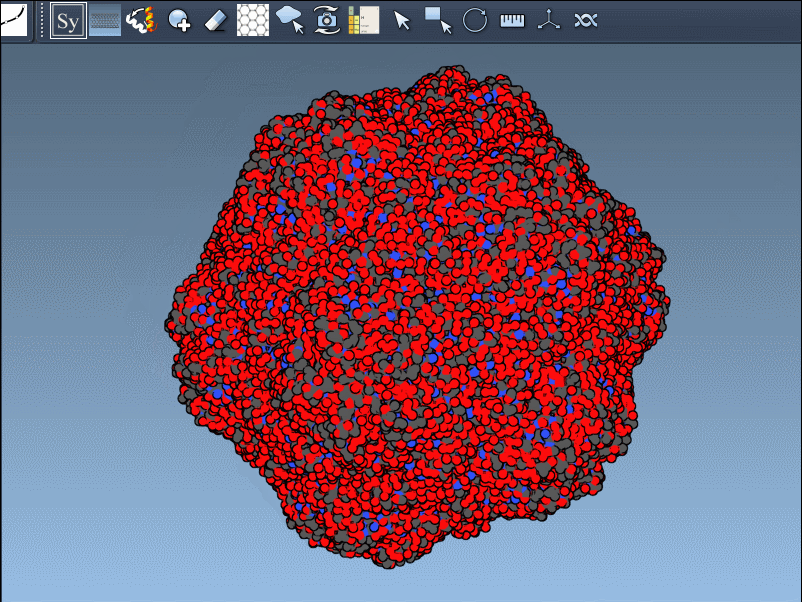
Once the Symmetry Mate Editor is activated, you’ll see small control nodes in the viewport that represent symmetry transformation positions extracted from CRYST1 or BIOMT records in the PDB file.
What makes this especially intuitive is the real-time preview capability. Simply hover over a node to see a transparent replica of the protein at the corresponding symmetric position. This alone helps you quickly assess which transformations are relevant to your modeling question.
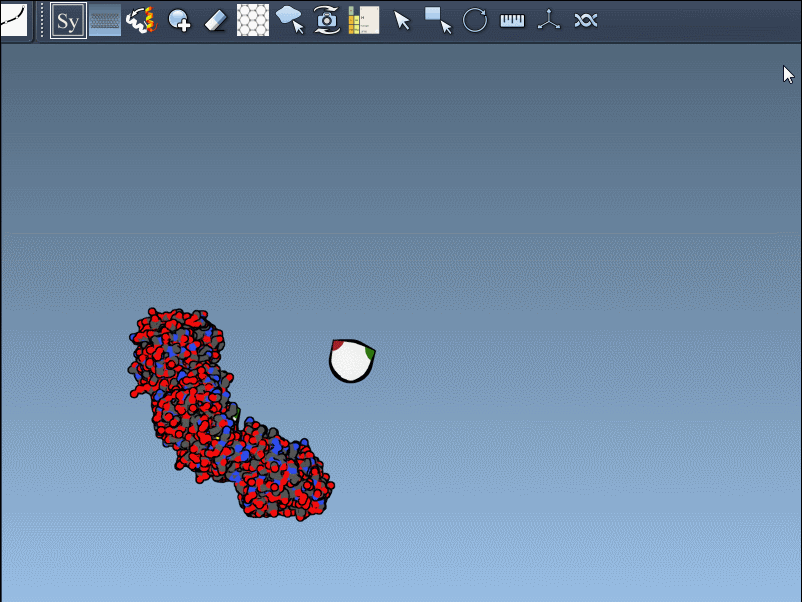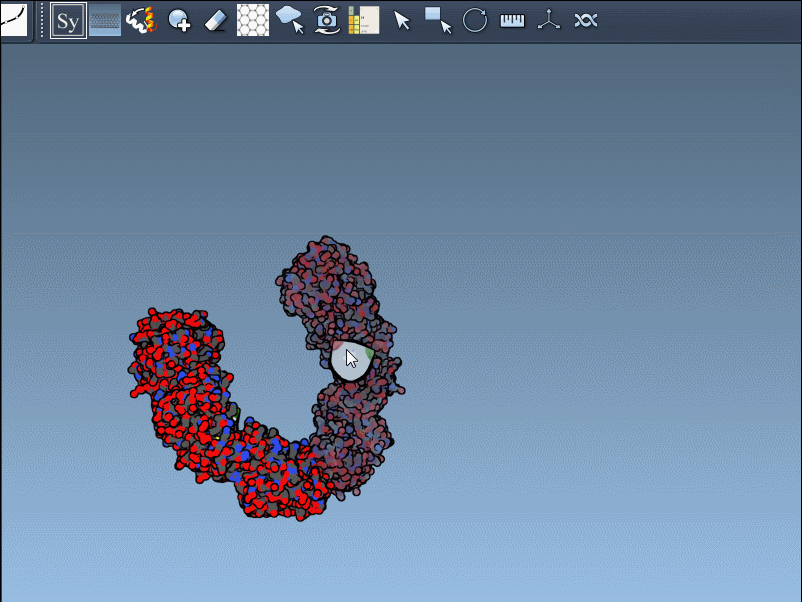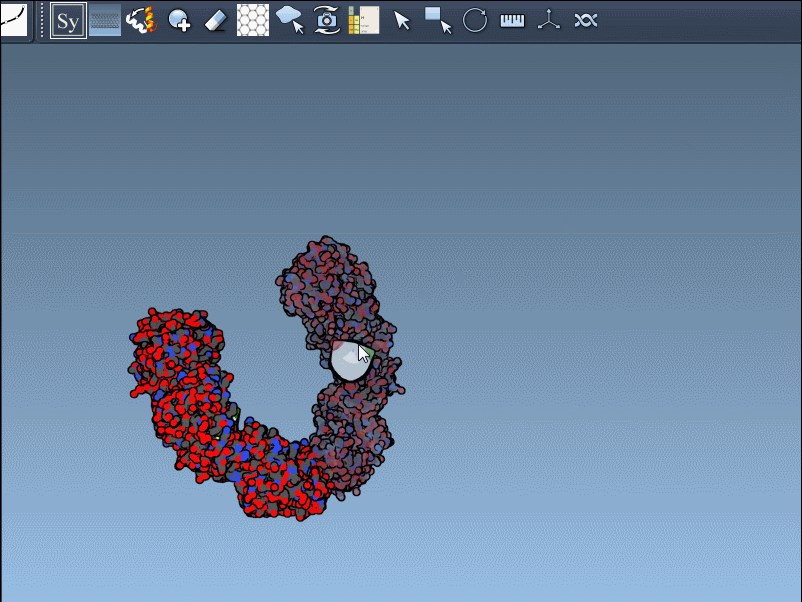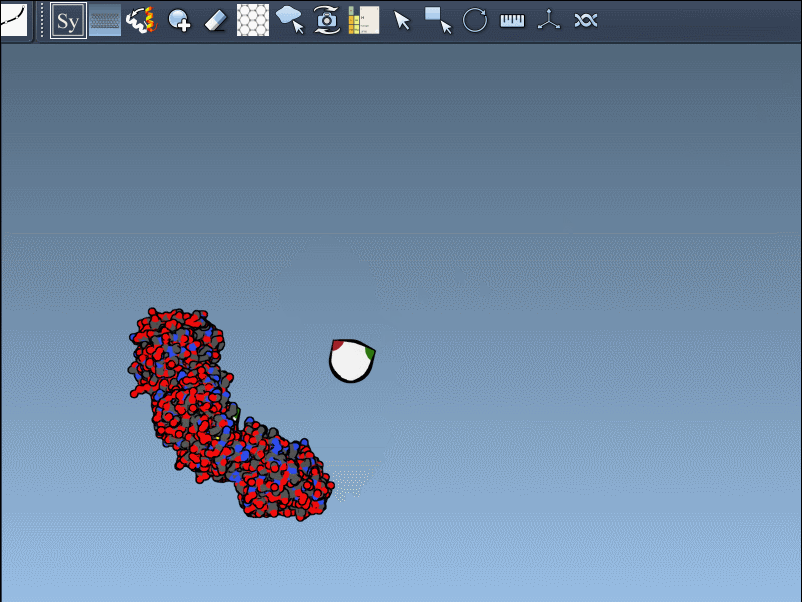
Left-Click to Confirm
If the previewed structure looks useful, left-clicking the control node will generate an actual replica at that position. You can repeat this for as many nodes as you’d like, selectively rebuilding the overall assembly without applying all transformations blindly.
Generating All Symmetry Mates at Once
If you already know you want all possible symmetry-related replicas generated, there’s also a one-step approach:
Hold Ctrl (or Cmd on macOS) and then hover and click a node.
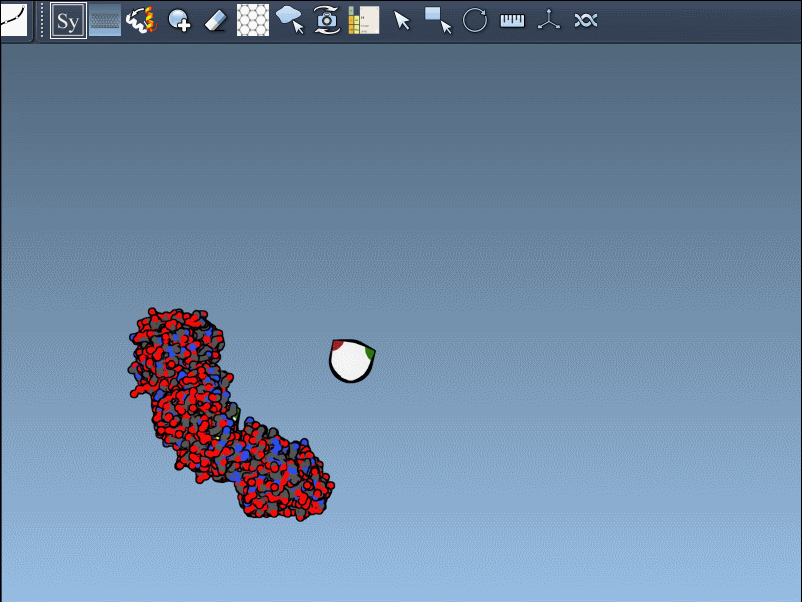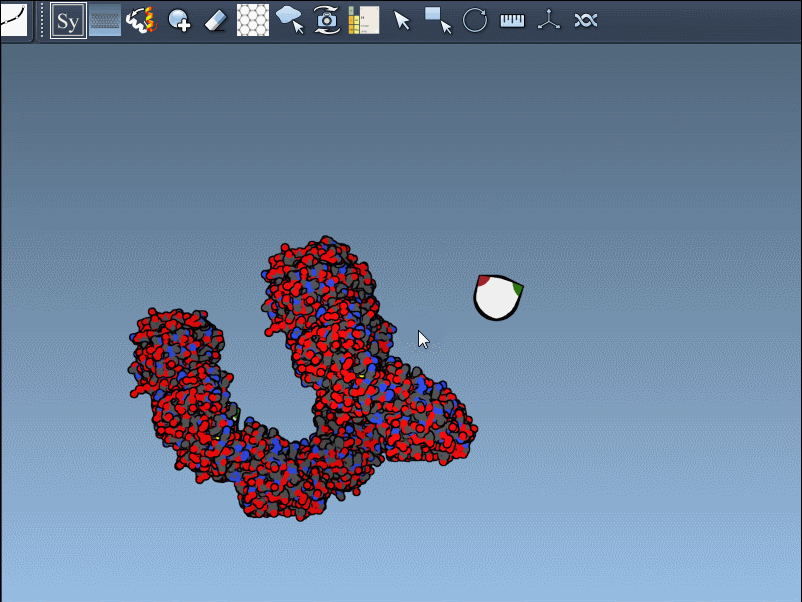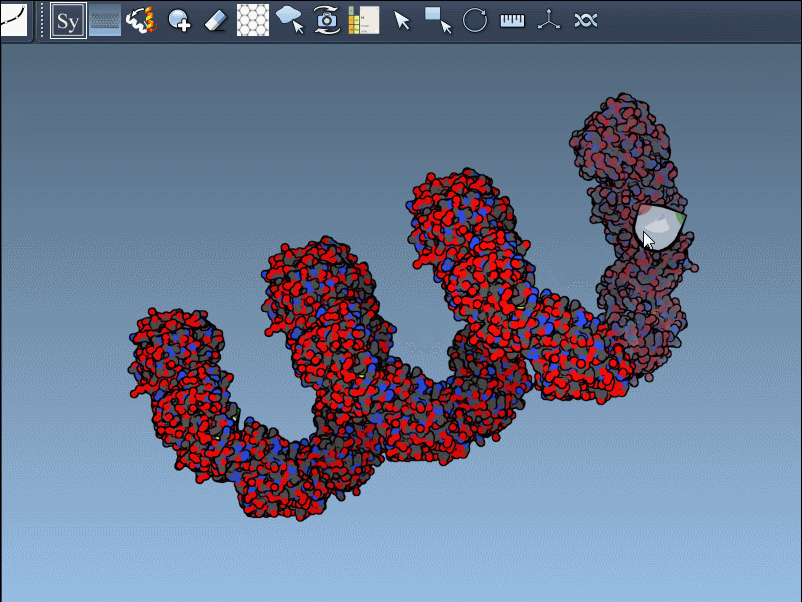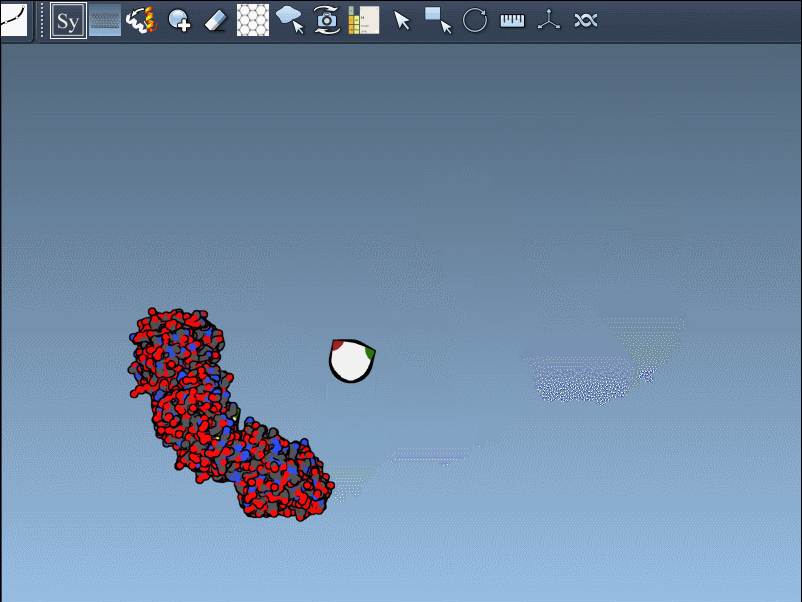
This rapidly previews and generates the entire biological assembly in one action — useful for reconstructing full oligomers or nanostructures.
Added Convenience: Undo, Scale, and Visual Control
You can:
- Undo any operation with Ctrl/Cmd + Z.
- Use Ctrl/Cmd + mouse wheel to zoom the density of control nodes by scale.
- Switch between CRYST1 and BIOMT symmetry sources via the editor’s interface.
This added flexibility allows you to maintain full control over how you reconstruct the assembly, combining automation with user validation.
Why This Matters for Molecular Modelers
Building out symmetry mates gives insight into key biological features like protein-protein interfaces, quaternary structure, and repeated binding sites. The real-time previews provide a clear and visual way to decide what to keep and what to ignore, greatly improving transparency in modeling workflows.
Especially if you’re planning simulations or designing symmetric systems (like cages or bundles), this feature integrates biological data with visual editing — helping you go from asymmetric unit to full assembly, quickly and reliably.
To learn more, visit the full documentation page: https://documentation.samson-connect.net/tutorials/symmetry/generating-symmetry-mates/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





