Many molecular modelers have faced the challenge of reconstructing complete protein assemblies from asymmetric crystallographic units. Knowing how subunits interact symmetrically is key for understanding biological function, designing protein-based materials, and setting up molecular simulations. But replicating these symmetric arrangements often feels like a tedious task, especially when time is limited and several configurations need to be explored.
If you’re using SAMSON, there’s a convenient and visual way to uncover these interactions using the Symmetry Mate Editor. In this post, we’ll walk through how to preview and generate all symmetry mates at once, based on transformation data directly embedded in PDB files.
Why previewing all replicas matters
Usually, one can click individual control nodes to generate protein replicas manually. But what if you want to instantly visualize the complete biological assembly? Or ensure that you’re generating all relevant symmetry mates that surround a unit in a crystal lattice?
Instead of clicking nodes one by one, SAMSON lets you preview (and create) all symmetry-generated replicas with a single modified click—saving time and making it much easier to identify quaternary structures and repeat patterns at a glance.
How it works
Once you’ve loaded a PDB file that contains transformation records (CRYST1 or BIOMT), follow these simple steps in the Symmetry Mate Editor:
- Activate the Symmetry Mate Editor: Press
Shift + Eand search for it, or find it under the left-side viewport menu: … > General > Symmetry Mate Editor. - The viewport will then show control nodes representing symmetry operations.
- Hold
Ctrl(on Windows/Linux) orCmd(on macOS). - While holding the key, hover over any control node to preview all symmetry mates in real time.
- Still holding the key, left-click to generate all replicas at once.
This technique works seamlessly with any structure containing either CRYST1 or BIOMT transformation data, and you’ll see a full set of symmetric chains appear in a single action, color-coded by transformation type (white for CRYST1, yellow for BIOMT).
Visual example
Below is an illustration of this feature using symmetrical protein data (in this case, forming an icosahedral structure):
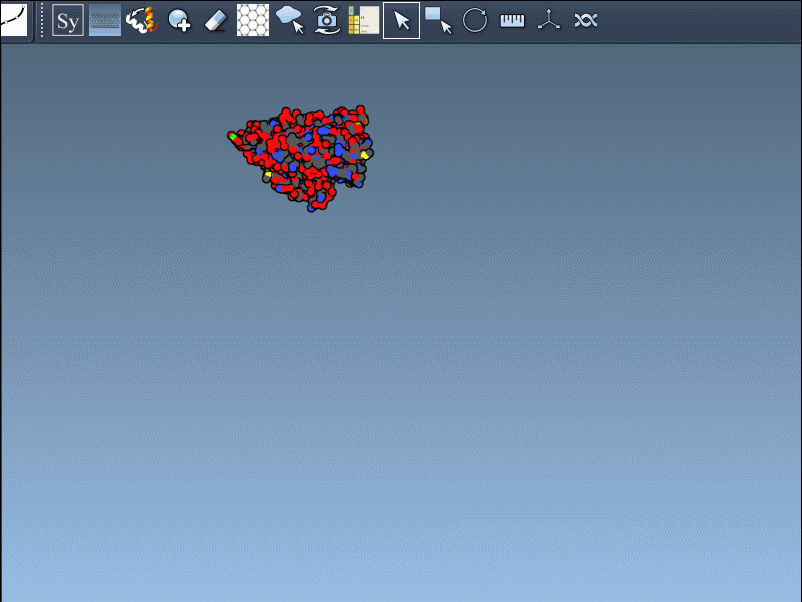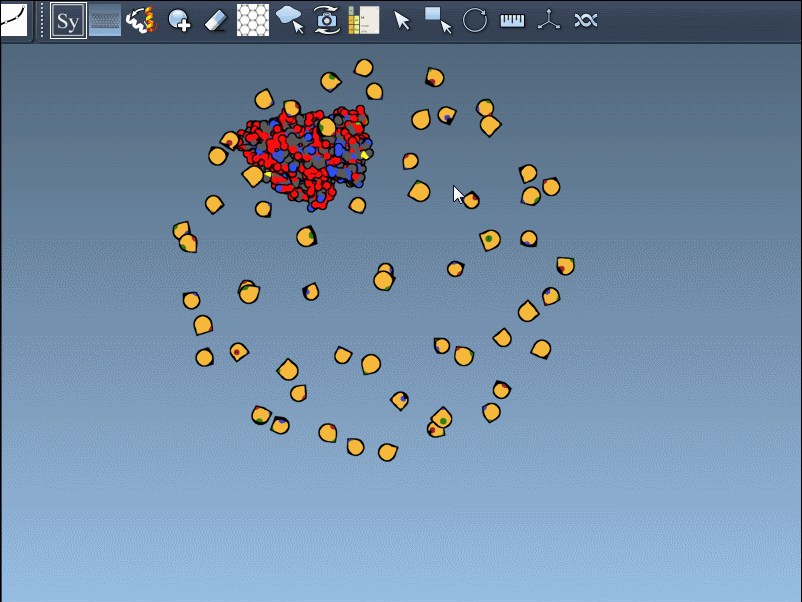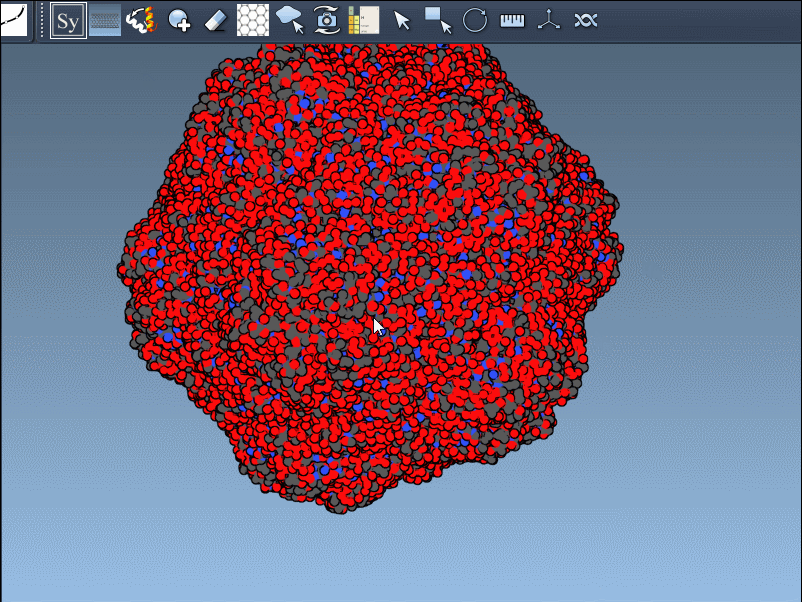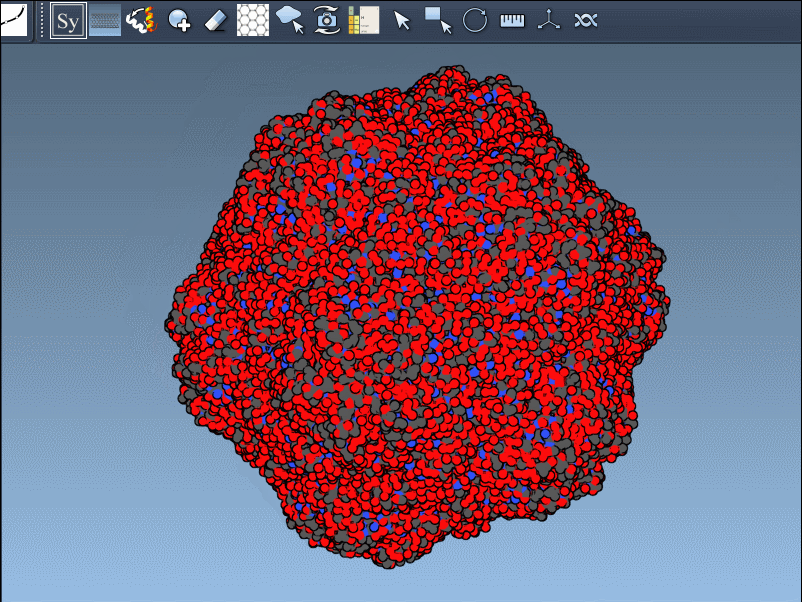
This becomes especially useful in scenarios like designing symmetric cages, evaluating interface repetition across subunits, or setting up simulation boxes that include full biological assemblies.
Undoing replicas
Don’t worry about making permanent changes — you can always return to the previous state using Ctrl + Z (or Cmd + Z). Each replica can be removed via the undo command, keeping your design process flexible.
Final notes
The ability to preview and generate all symmetry mates at once represents a small but impactful improvement for researchers exploring molecular assemblies. It’s especially helpful when dealing with complex or highly symmetric systems, where visualizing the whole picture early on can inform and accelerate design decisions.
To learn more about symmetry mates and use other powerful visualization tools built into the Symmetry Mate Editor, visit the full documentation:
https://documentation.samson-connect.net/tutorials/symmetry/generating-symmetry-mates/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON from https://www.samson-connect.net.





