One of the most common challenges in molecular modeling today is exploring how small, strategic changes in chemical structures can affect properties like binding affinity or biological activity. If you’ve ever wanted a fast and visual way to generate and test analogs of a molecule—without scripting or complicated workflows—the SMILES Manager in SAMSON offers an accessible and intuitive solution.
In this post, we focus on a very specific and practical feature: replacing patterns in molecules using SMARTS. This approach can help chemists, modelers, and drug designers better understand structure-activity relationships by performing a method akin to Positional Analogue Scanning (PAS) within a few clicks.
What’s the idea?
Using the SAMSON SMILES Manager, you can search for a molecular pattern—such as an aromatic proton or functional group—and either replace it or attach a new atom or group. For example, you might want to know what happens when an aromatic carbon with a hydrogen ([cH]) is replaced by nitrogen (N), or what new interactions arise when you attach a fluorine (F).
How it works
The SMILES Manager interface guides you through the process in four simple steps:
- Start from a single molecule: Paste in a SMILES code or select a molecule in your SAMSON document.
- Define the pattern: Use SMARTS notation to specify what to look for. For instance,
[cH]highlights aromatic hydrogens—ideal for slight, rational modifications. - Replace or attach: Swap out the identified pattern with another atom or group of your choice.
- Run the scan: Click a button to generate your analogs. 2D depictions and SMILES codes appear instantly in the results panel.
This makes it effortless to generate a focused set of analogs for further study. Instead of manually altering structures or writing custom scripts, you can probe your chemical hypothesis using a graphical workflow.
A visual walkthrough
Here’s how the pattern selection and replacement works visually:

In this example, we highlight all the positions in the molecule corresponding to the [cH] SMARTS pattern.
After defining the desired replacements, clicking “Run” produces a panel like the one below:
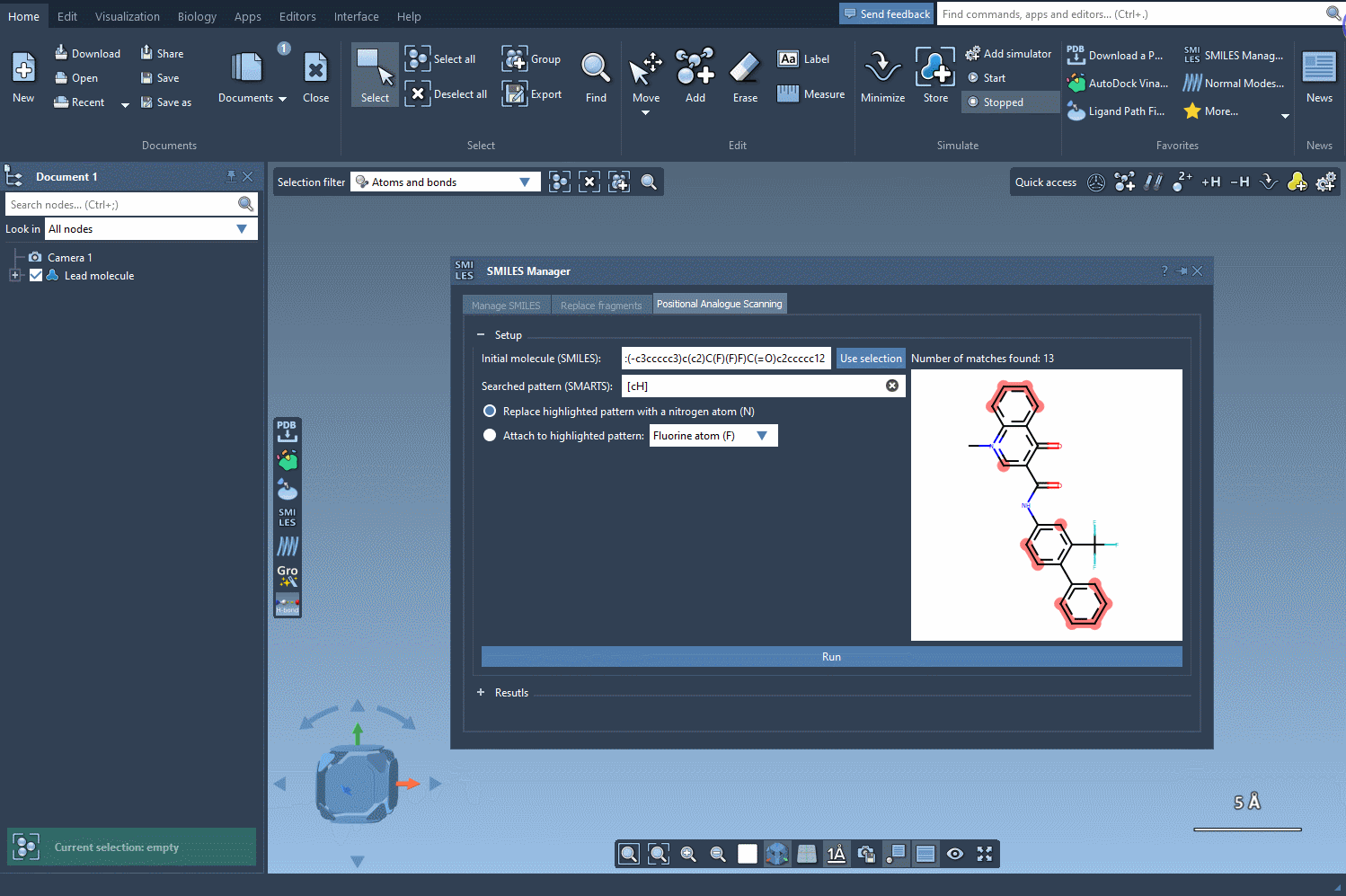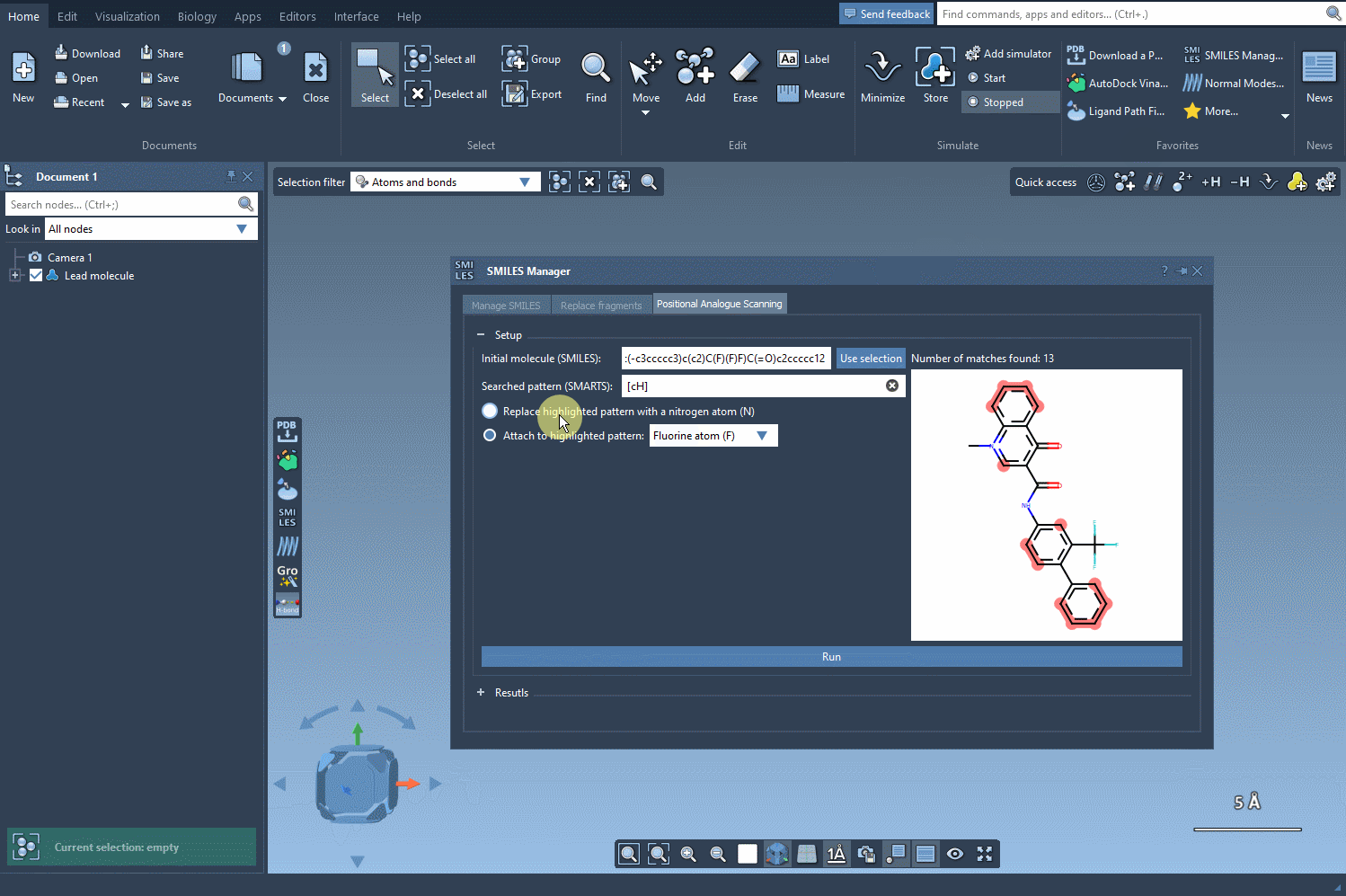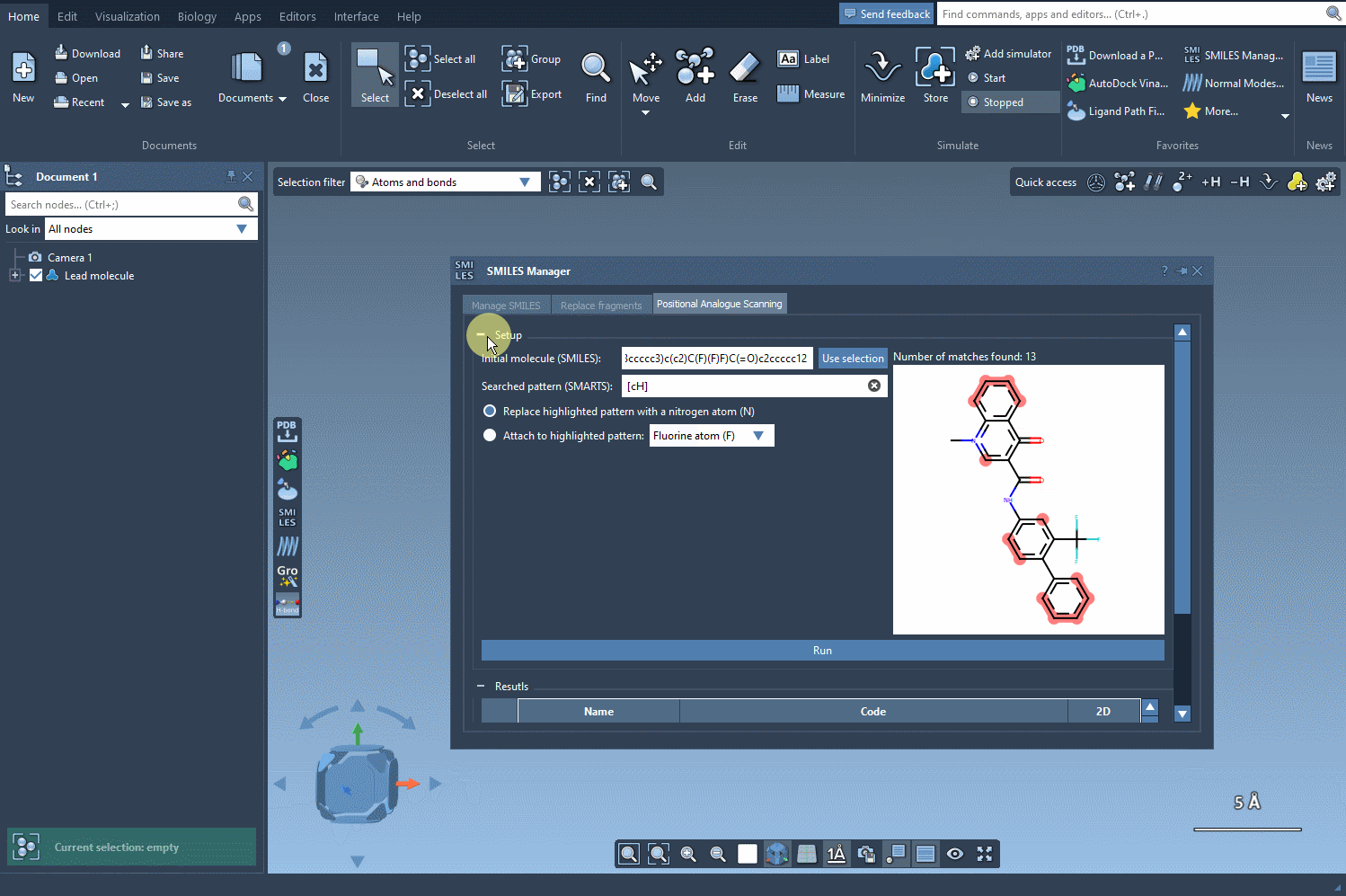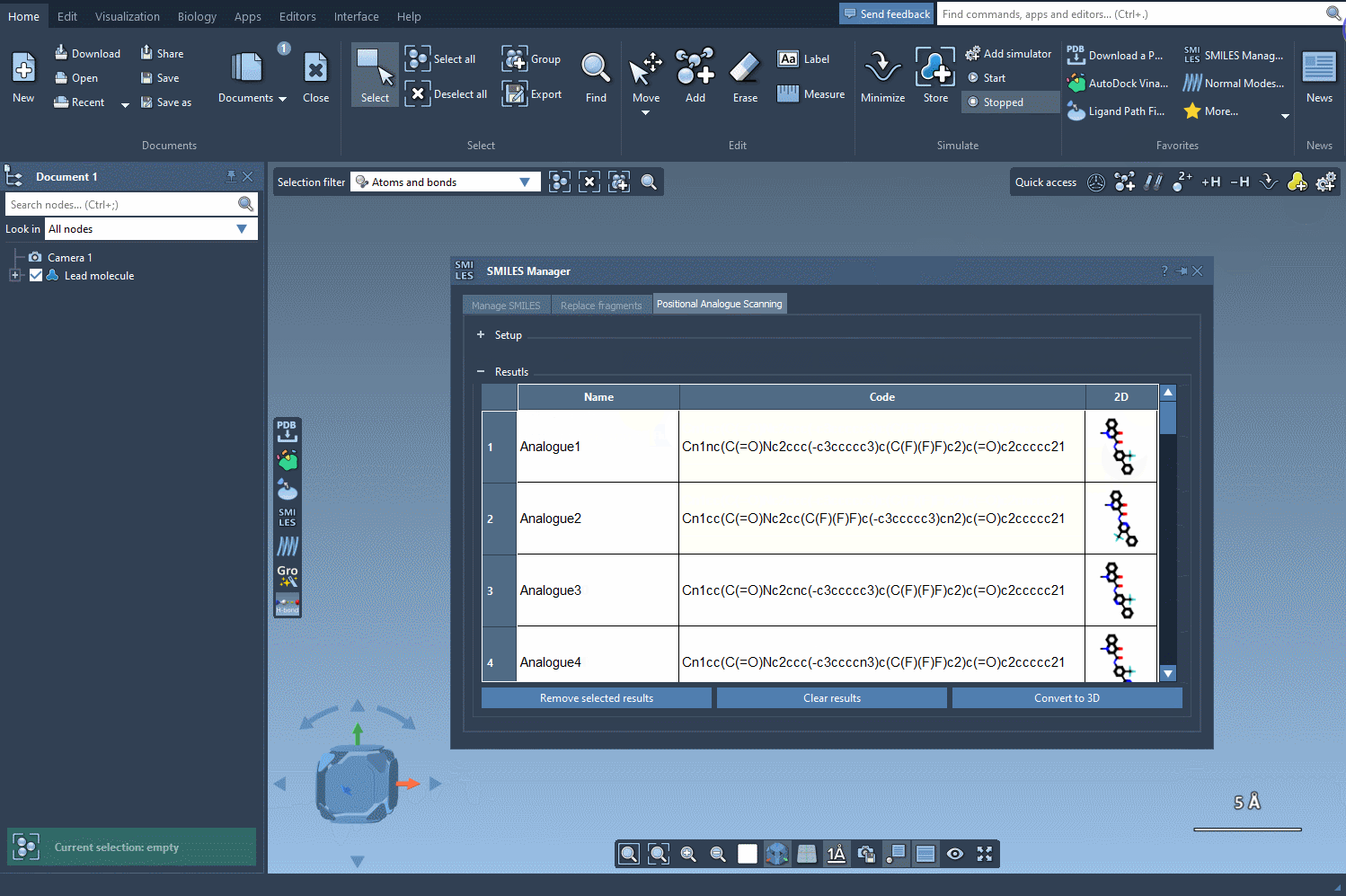
Each variant can be visualized, modified, or exported for downstream analysis.
Why it matters
One of the biggest barriers to systematic analog generation is convenience. Researchers often fall back on limited manual edits or tedious automation steps. The SMILES Manager serves as a bridge between intuitive graphical control and powerful chemical transformation rules.
Whether you’re designing focused libraries for virtual screening or just curious about a single substitution’s effect on activity, this tool brings efficiency and transparency.
Want to customize further?
The interface allows direct modification of generated analogs (including renaming and SMILES edits), 3D structure generation with one click, and even integration with other tools like Autodock Vina Extended for docking experiments—all from the same panel.
Learn more by reading the full tutorial.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





