When working on optimizing small molecules for better binding affinity or selectivity, one often faces the question: What happens if I replace or attach something at this position? Trial-and-error can be time-consuming, and manual modifications in 3D editors are prone to error and inconsistency. Fortunately, the SMILES Manager extension in SAMSON offers a fast and intuitive way to tackle this challenge using pattern-based analogue generation.
This technique allows you to define a chemical pattern to look for in your molecule (like aromatic hydrogens) and automatically suggest variants by replacing or attaching specified groups. It drastically reduces the manual work required to generate analogs, while offering flexibility in defining the scope of your exploration.
Step-by-step: Modify Your Molecule at Specific Patterns
Start by loading your molecule into SAMSON, either by pasting its SMILES code or interacting directly with the structure in your SAMSON workspace. From there, using the SMILES Manager interface, you can define the substructure pattern to scan using SMARTS syntax. For example, entering [cH] will target hydrogen-bearing aromatic carbon atoms.

Once these positions are identified, you can choose what kind of modification you’d like to perform:
- Replace with a different atom, such as nitrogen (N)
- Attach a functional group — e.g., fluorine (F) or methyl (CH3)
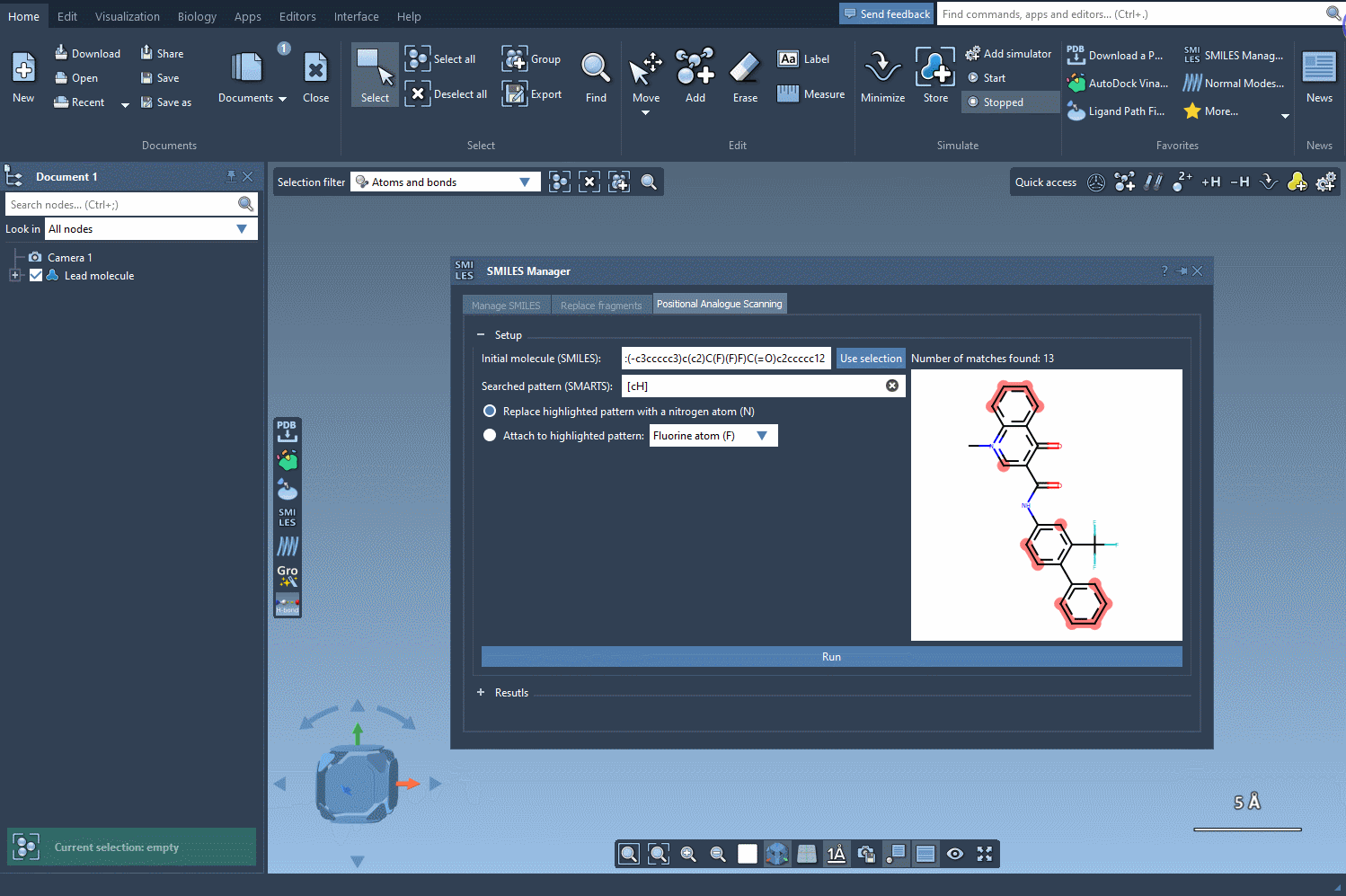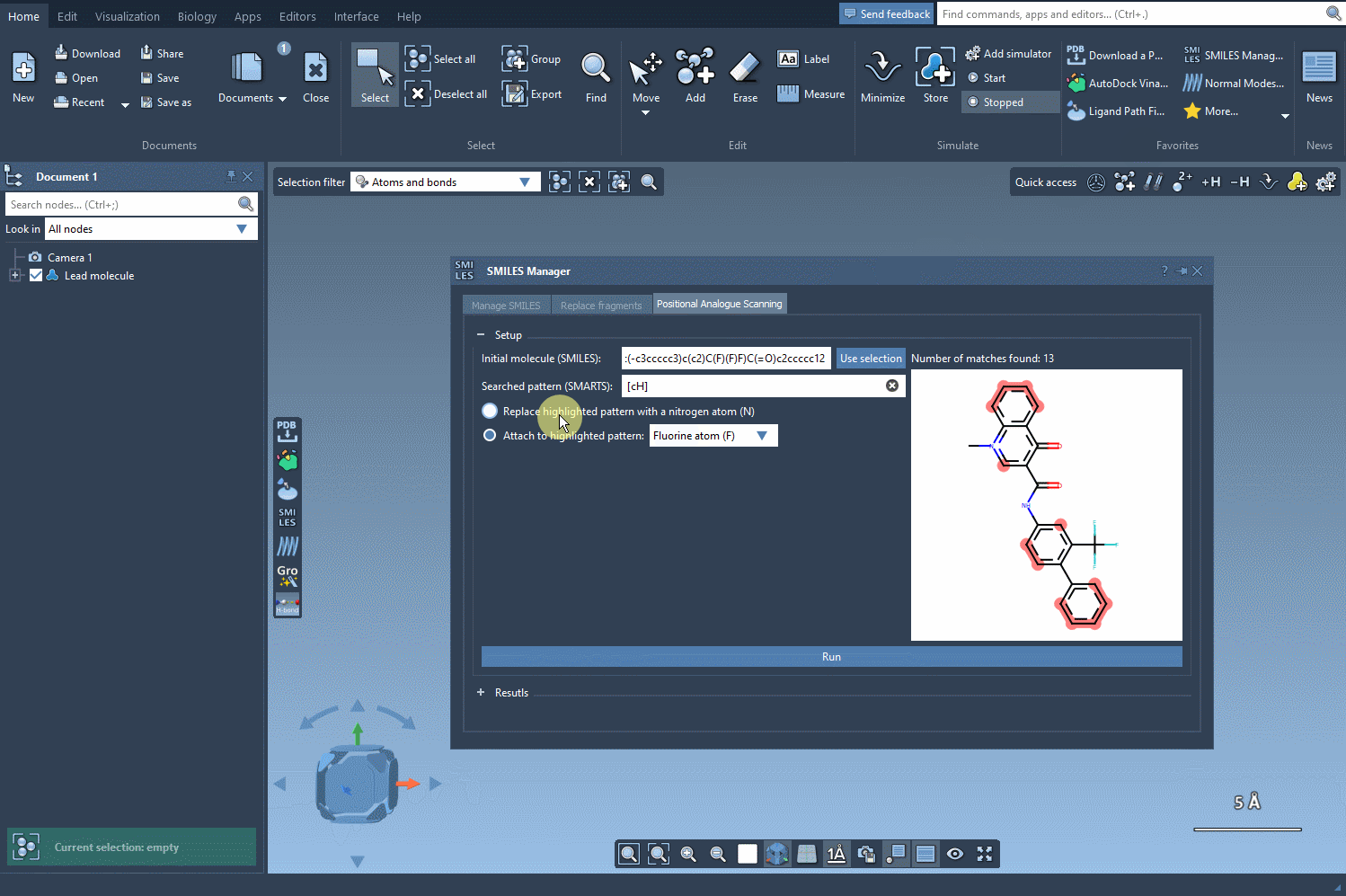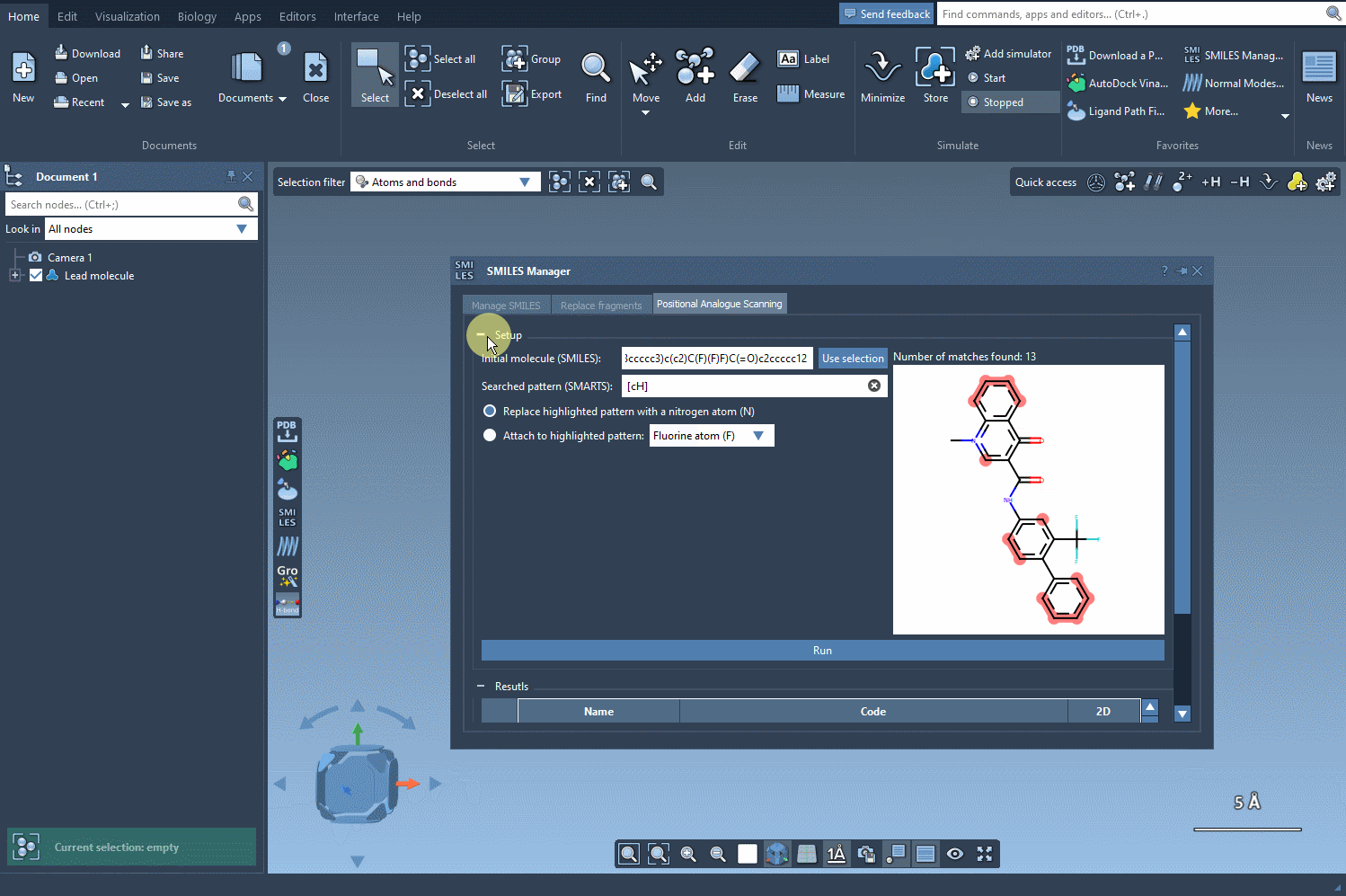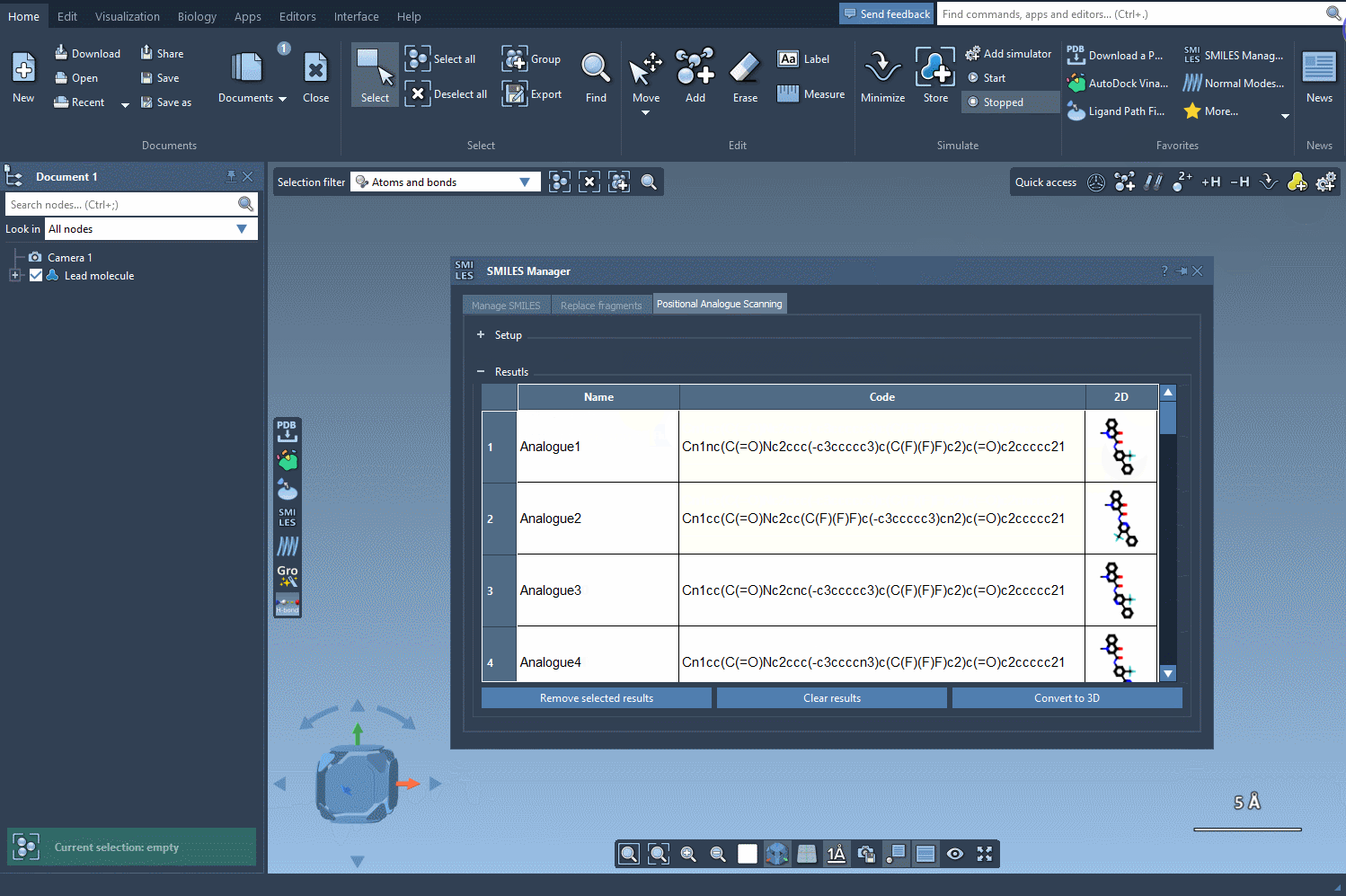
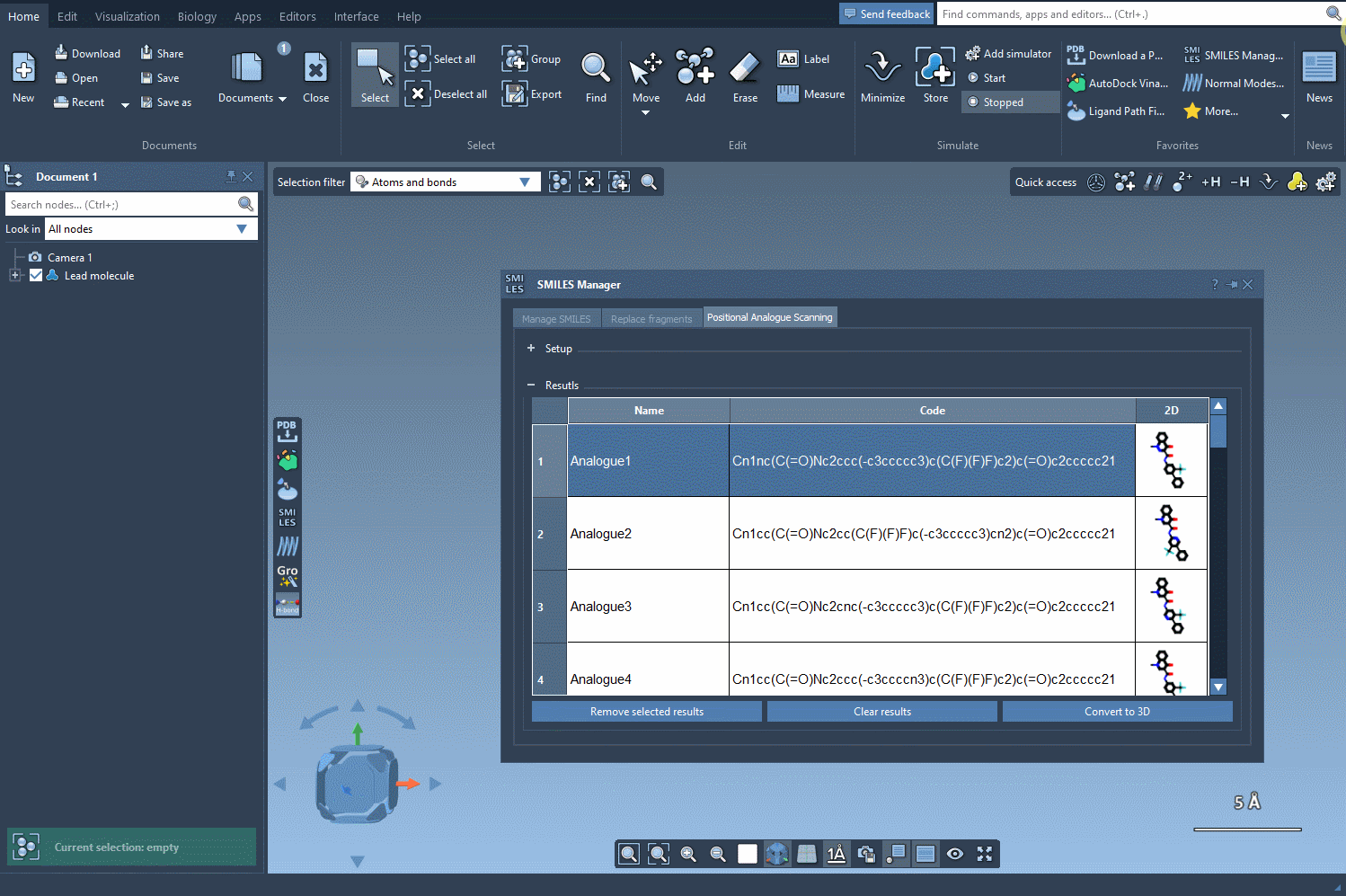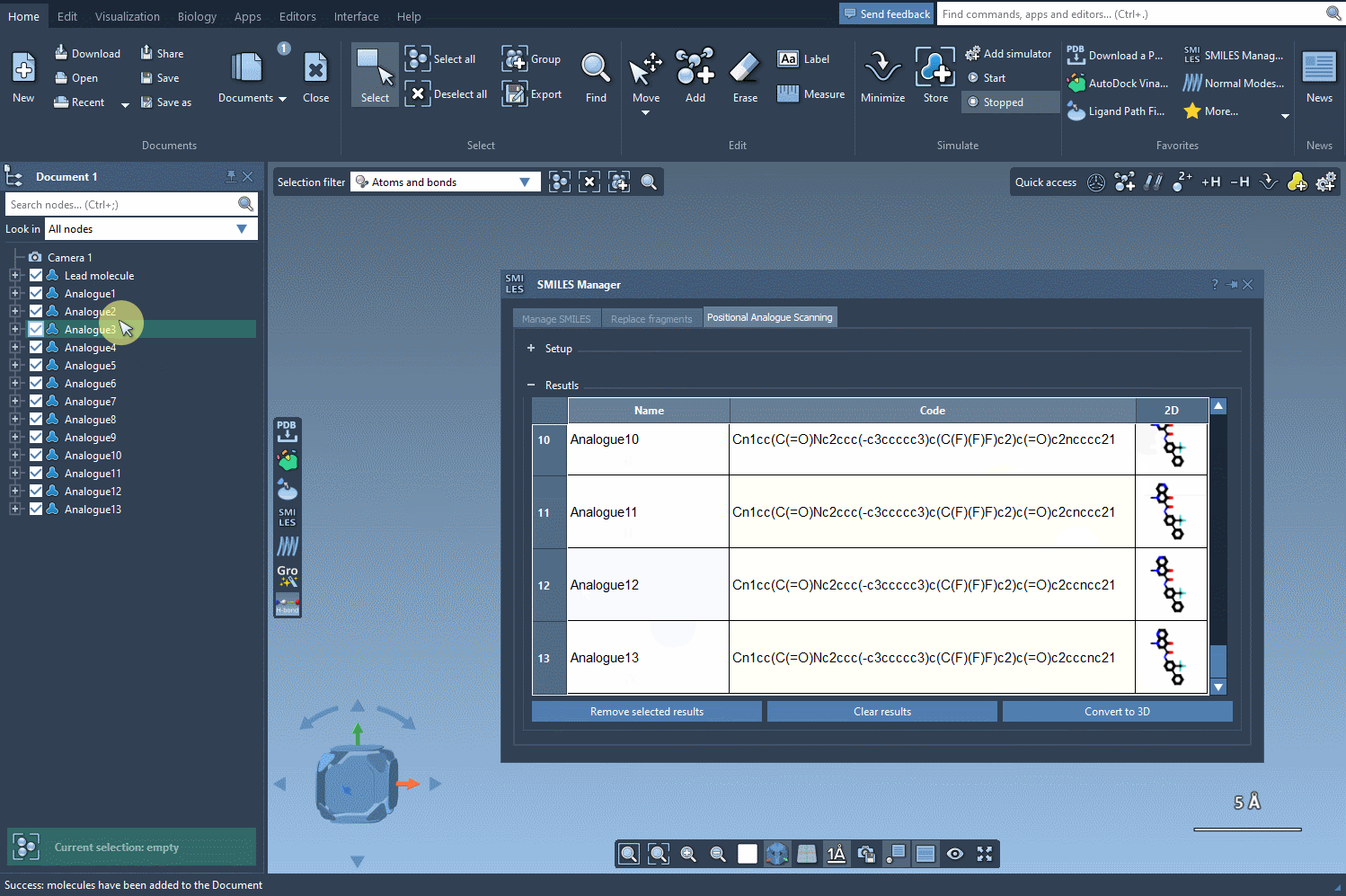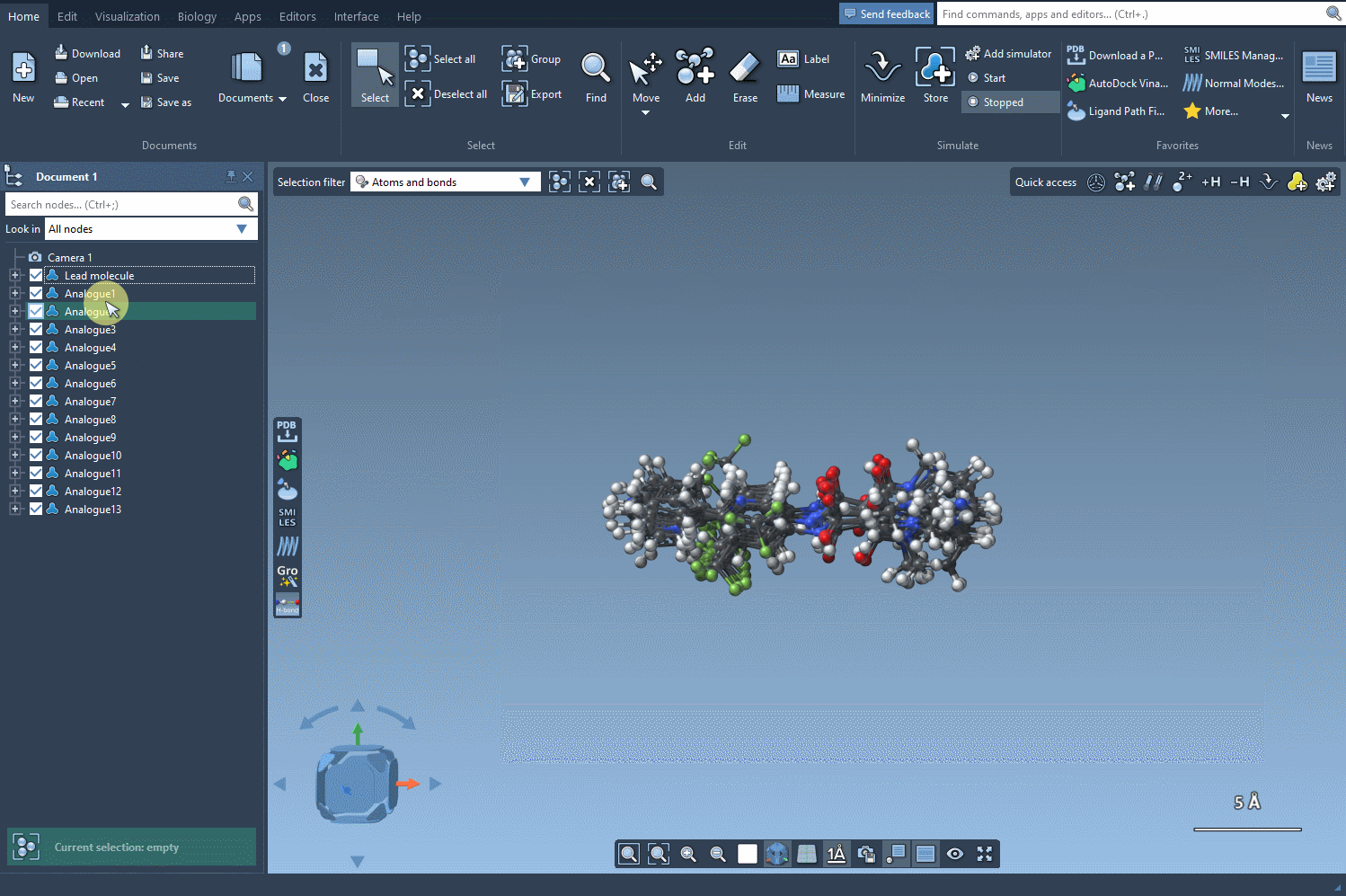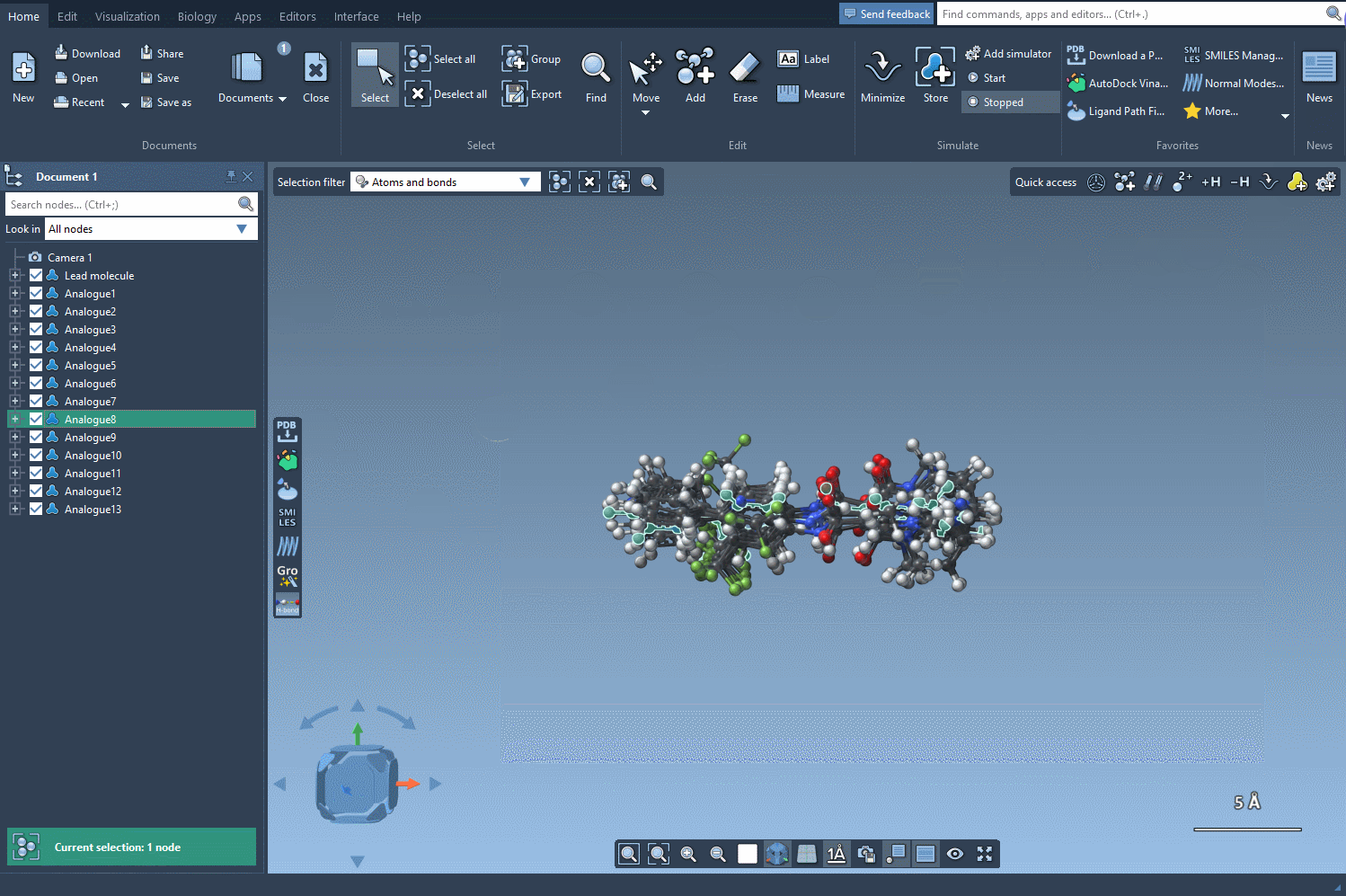
The SMILES Manager will then automatically generate a list of analogs, showing their SMILES code and 2D depictions in a convenient table. All of this happens almost instantly.
Modify, Organize, and Export Your Analogs
The built-in results table makes managing your generated molecules easy. Need to rename an analog? Just double-click its name. Want to make further manual edits? You can adjust the SMILES directly within the table. You can show/hide 2D depictions, or even selectively remove entries you no longer need.
Each analog can also be converted into its 3D structure with a simple right-click, allowing you to proceed immediately to further analysis, such as docking or visualization in 3D.
Why Use This Approach?
If you’re trying to systematically explore the impact of substituents or slightly varied scaffolds, this method is especially helpful. Rather than manually drawing and pre-optimizing molecules elsewhere before import, you can:
- Automatically generate consistent, chemically meaningful analogs
- Avoid transcription errors and wasted time on manual drawing tools
- Integrate pattern selection and modification in one place
It’s a practical method for lead optimization campaigns, SAR studies, or quick iterations based on docking results. Plus, using SMARTS gives you full control over defining where changes should happen — no black boxes.
To learn more, visit the full documentation page here: Perform Positional Analogue Scanning using the SMILES Manager.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





