If you’ve ever loaded a PDB file into your molecular modeling environment only to find a lone asymmetric unit floating in empty space, you’re not alone. Most PDB files describe just the minimal repeating unit of a biological assembly or crystal lattice. But for functional analysis, interface exploration, or simply seeing the whole picture, visualizing the full symmetric assembly is essential.
This post introduces the Symmetry Mate Editor in SAMSON, a convenient way to explore and recreate symmetric copies of proteins directly from built-in transformation data (like CRYST1 and BIOMT records) in the PDB file — without scripts, manual transformations or external tools.
Why Seeing Symmetry Matters
- Understand how a monomer organizes into biologically active multimers.
- Find potential protein-protein interfaces that are invisible in the asymmetric unit.
- Reveal how ligand binding or allosteric effects might propagate across quaternary structure interfaces.
- Generate realistic full complexes for simulation and design workflows.
Straightforward Access to Symmetry Mates
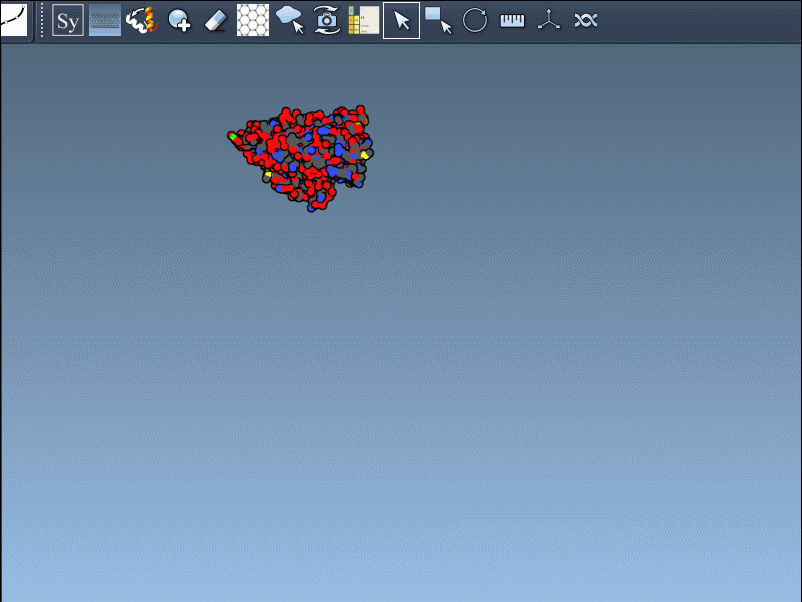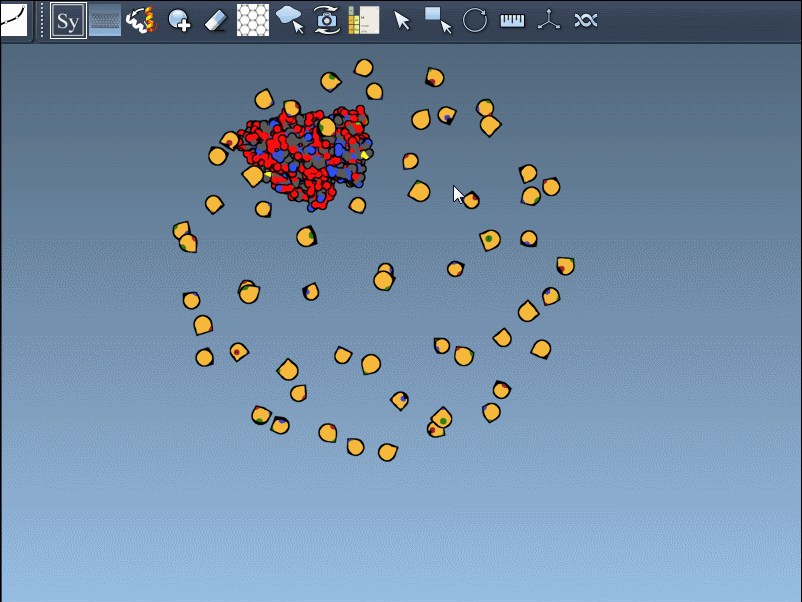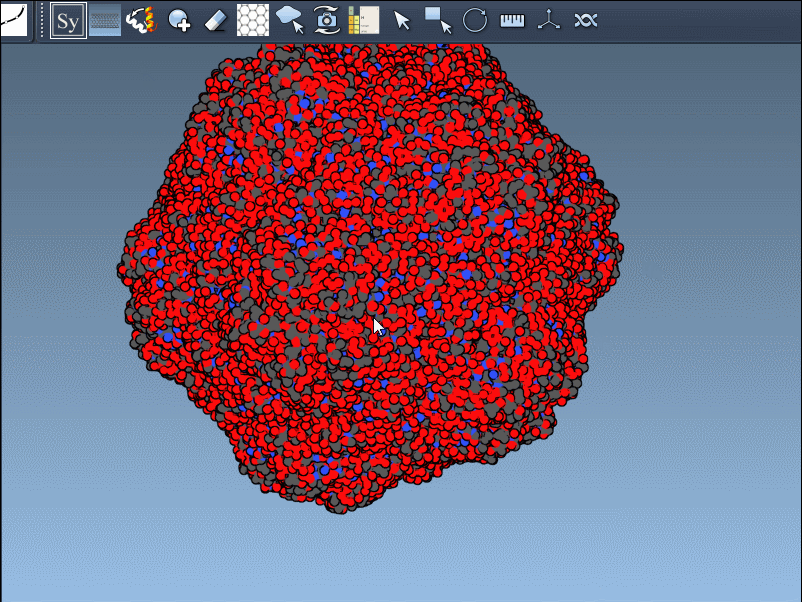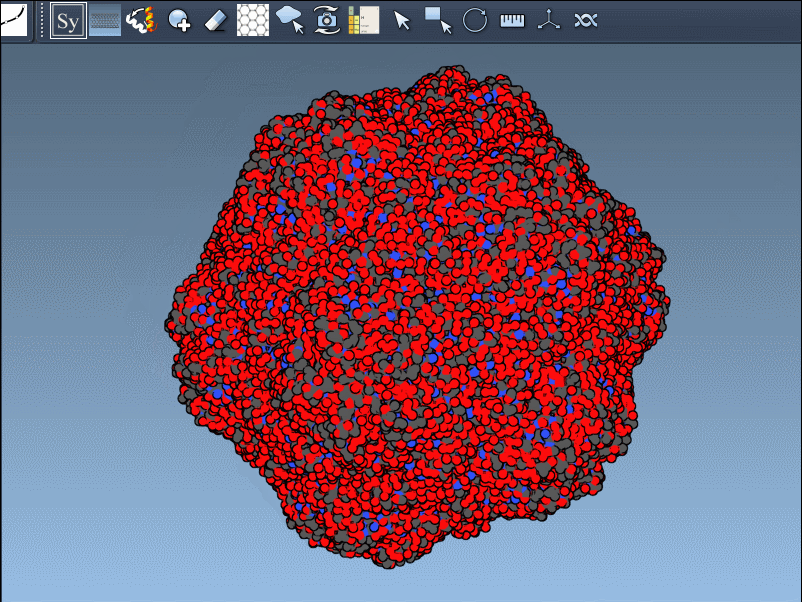
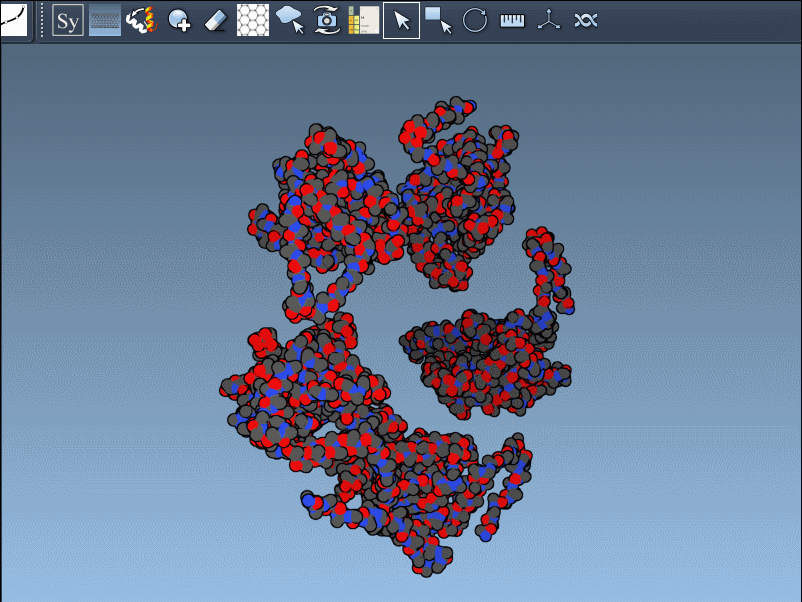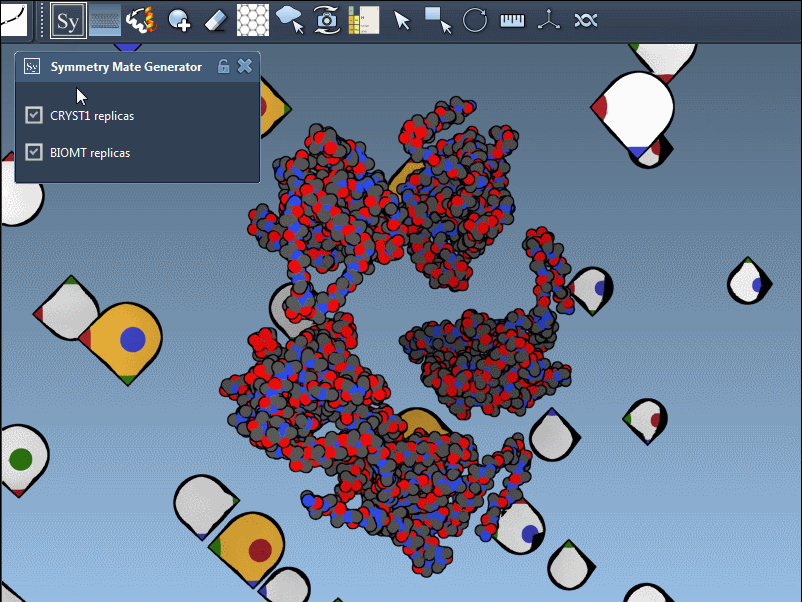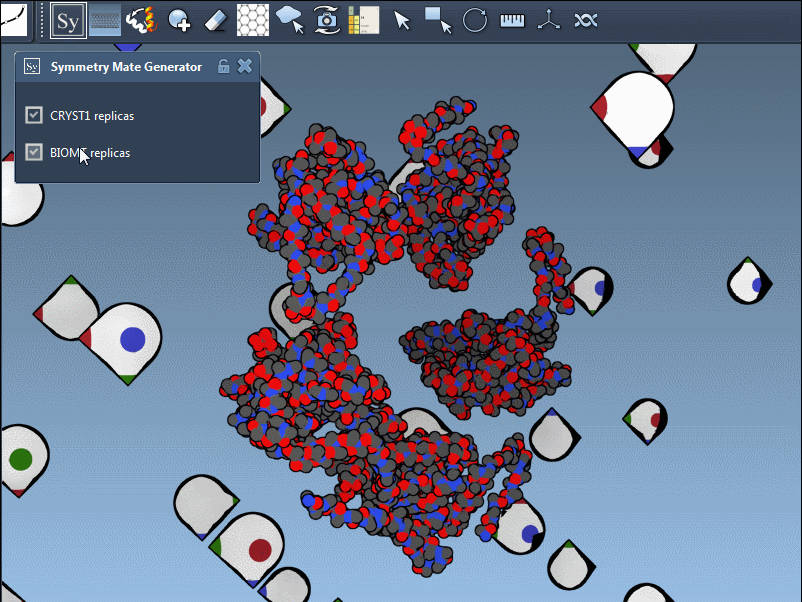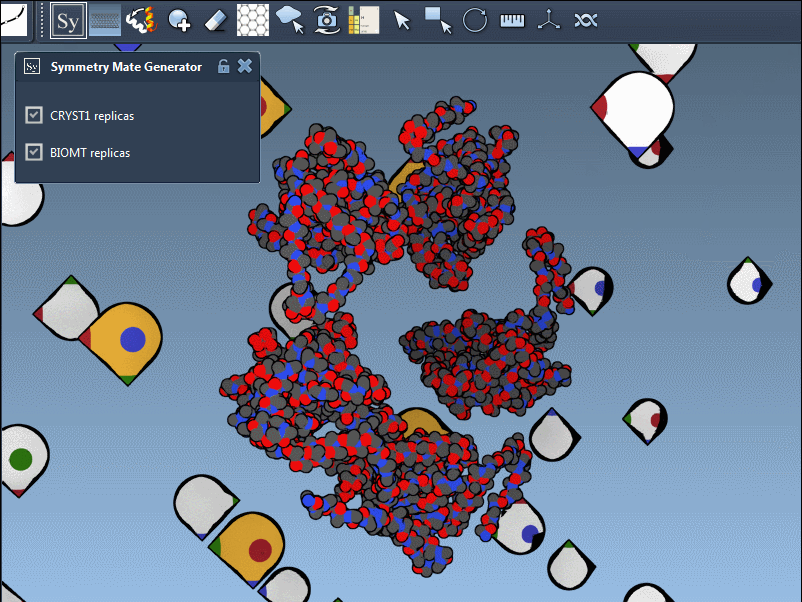
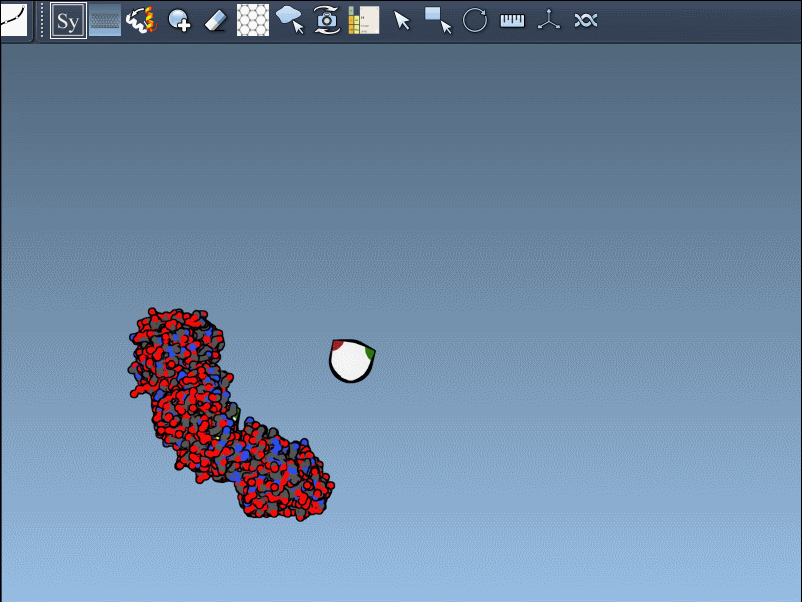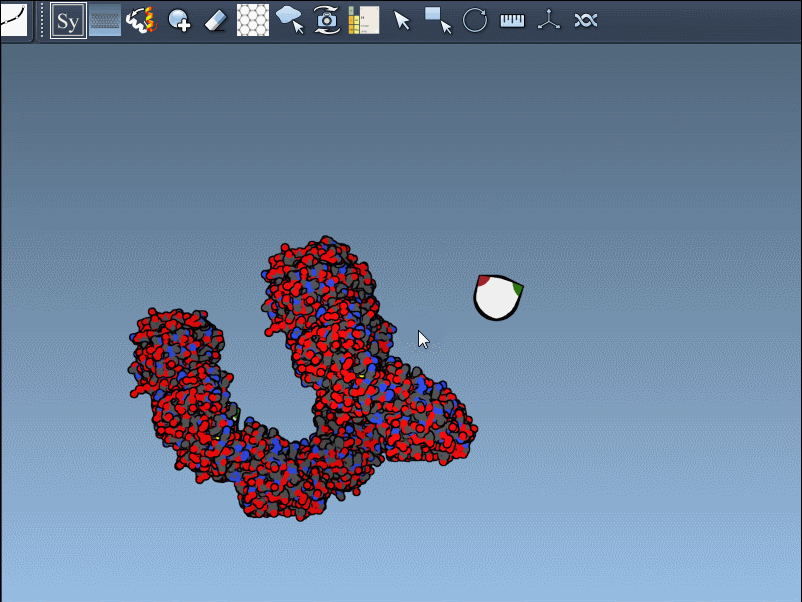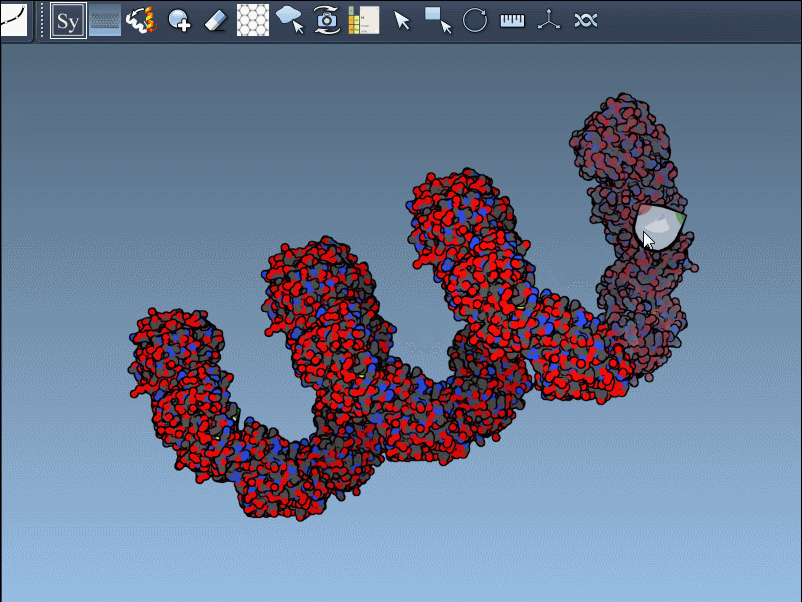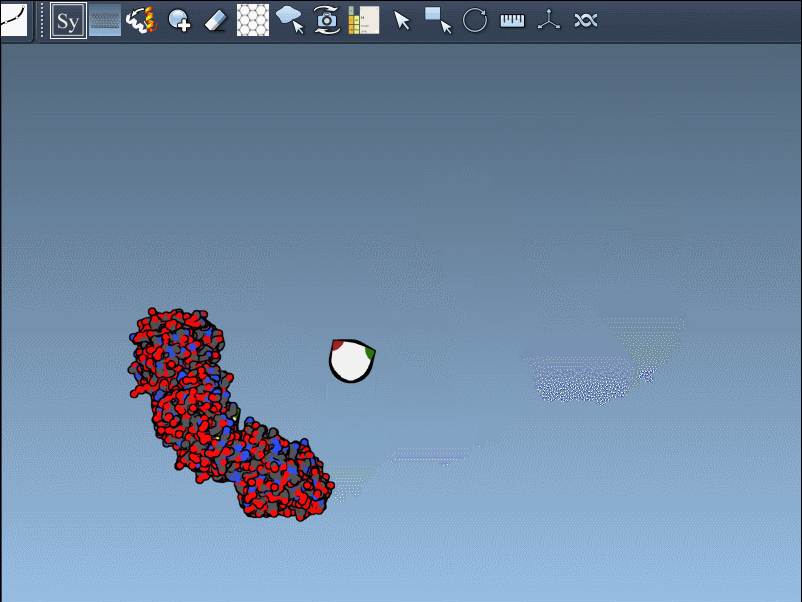
Once you’ve added and activated the Symmetry Mate Editor in SAMSON, control nodes appear around your structure, each representing a symmetry transformation from either the crystal lattice (CRYST1) or biological assembly (BIOMT) data.
Hover a node to preview the symmetric copy in real-time.
Left-click the node to make the copy permanent.
Press and hold Ctrl (or Cmd) while hovering and clicking to generate all symmetry mates in one shot.
CRYST1 vs. BIOMT – Know What You’re Looking At
The editor distinguishes between two types of symmetry data:
- CRYST1 transformations (white control nodes) reflect how molecules pack in a crystal.
- BIOMT transformations (yellow nodes) relate to biological assemblies curated by PDB authors.
You can choose which transformation type to explore based on your modeling goals. For biomolecular simulations or interface studies, BIOMT assemblies are typically more relevant.
Extra Control While Modeling
The Symmetry Mate Editor gives you fine control:
- Use Ctrl/Cmd + scroll wheel to scale the number of visible transformation nodes.
- Undo any replica generation with Ctrl/Cmd + Z.
- Use color-coded chain ribbons to easily distinguish parts of the complex.
Tips and Use Cases
If you’re designing oligomeric protein cages or want to simulate symmetric interfaces, symmetry mates are a shortcut to accurate structural context. Want to analyze repeated binding sites or compute RMSDs across chains? Combine this with the Symmetry Detection extension.
This fast visual feedback loop shortens exploratory workflows and minimizes manual setup.
For more details and step-by-step usage, visit the full documentation page: https://documentation.samson-connect.net/tutorials/symmetry/generating-symmetry-mates/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON for free at https://www.samson-connect.net.





