Whether you’re building a homology model, refining a predicted structure, or preparing a system for simulation, strained backbone conformations can introduce problems. Outliers in φ (phi) and ψ (psi) backbone dihedral angles may go unnoticed in a 3D viewer, but lead to unrealistic behaviors during simulations. Fortunately, SAMSON’s Interactive Ramachandran Plot Extension offers a direct and intuitive way to detect and edit these conformations at the residue level.
Why It Matters
Simulation-ready structures depend on chemical realism. Dihedral angle outliers in the Ramachandran plot are often the result of model building tools or structure prediction methods pushing local conformations into energetically disallowed regions. This results in steric clashes or unstable dynamics down the line.
The Interactive Ramachandran Plot Extension lets you correct these issues in a visual, real-time workflow. You don’t need to write scripts or dig into coordinate files — just select, drag, and refine.
Start by Loading a Protein
After installing the Interactive Ramachandran Plot Extension from SAMSON Connect, open SAMSON and fetch a protein structure using its PDB ID — for example, 1YRF. This can be done through Home > Fetch.
Visualize and Interact with the Ramachandran Plot
Open the app from Home > Apps > Biology > Ramachandran Plot. Click the Update button to generate a map of all backbone dihedral angles and identify outliers:
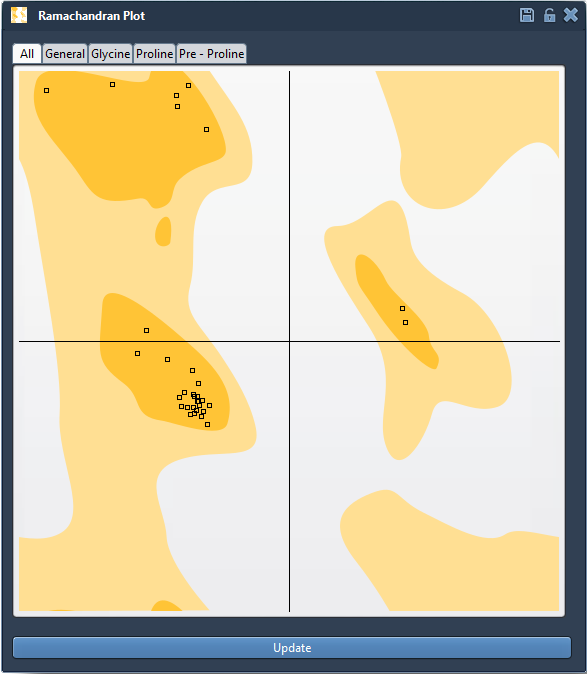
Residues appear as dots in region-dependent colors:
- Yellow areas – favored conformations
- White areas – energetically unfavorable
You can filter by residue type (general, glycine, proline, pre-proline) to focus on specific outlier categories.
Refine Conformations Directly
Option 1: Drag Points in the Plot
Click on a residue dot (e.g., a proline) to select it. The corresponding residue is highlighted in the 3D view, and dihedral angles appear in the status bar. You can then drag the point within the plot to adjust φ and ψ angles:

As you drag, the 3D structure updates instantly. If you’re not satisfied with the result, press Ctrl/Cmd + Z to undo.
Option 2: Use the Twister Editor
For more spatial control, activate the Twister editor from the left-hand menu in SAMSON. Twisting the structure this way also updates the Ramachandran plot in real time:
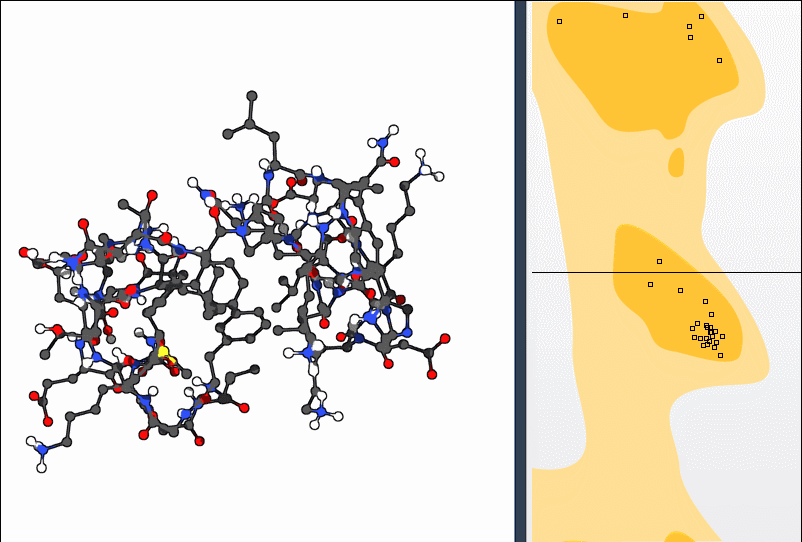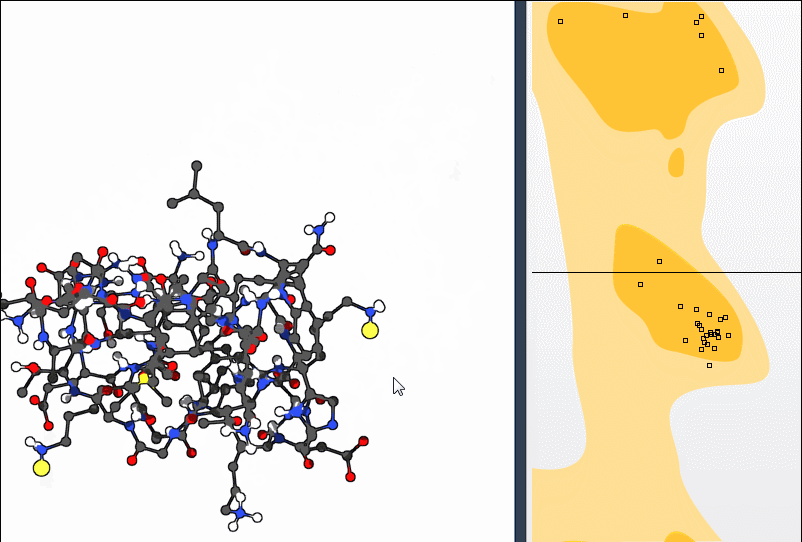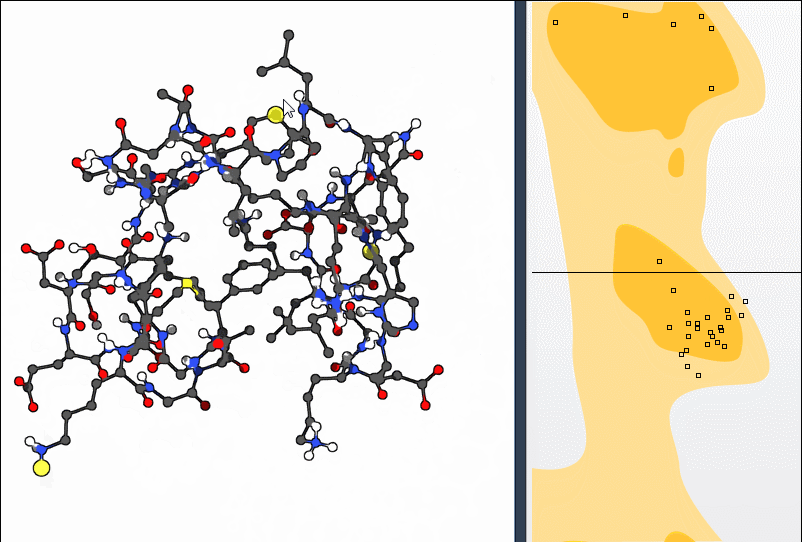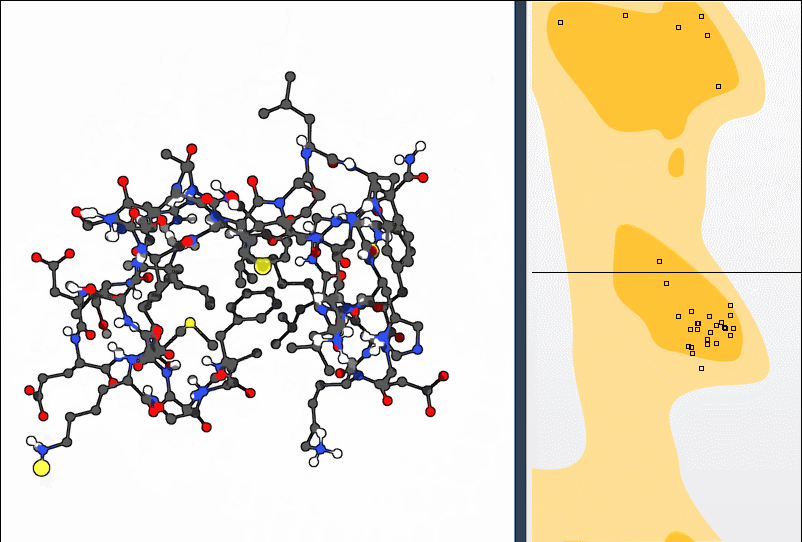
Combine with Normal Mode Analysis
If you’re using normal mode analysis (NMA) in SAMSON, you can combine global motion with local optimization. After applying a normal mode, use the Ramachandran plot to fine-tune residues toward favorable regions:
Wrap-up
This interactive editing approach strikes a balance between visual control and molecular insight — a useful addition to the toolkit of any structural modeler aiming for realistic and simulation-ready proteins.
For more details, see the full documentation: Interactive Ramachandran Plot Documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





