When working with structural protein data, it’s common to download a PDB file and start modeling or simulating right away. But sometimes what you see in the asymmetric unit isn’t the whole story. Many biological assemblies involve symmetric arrangements of subunits—that single monomer in your viewer might be just one piece of a much larger puzzle.
Recreating symmetry mates is an essential step in many workflows: it unveils the complete biological assembly, helps detect interfaces, explore potential binding sites, or construct realistic starting structures for molecular dynamics simulations.
Fortunately, in SAMSON, the Symmetry Mate Editor extension simplifies this process significantly. Instead of manually interpreting CRYST1 and BIOMT records or jumping between various tools, you can preview and generate symmetric protein replicas directly from the PDB data, interactively.
Visual Preview and Interactive Replication
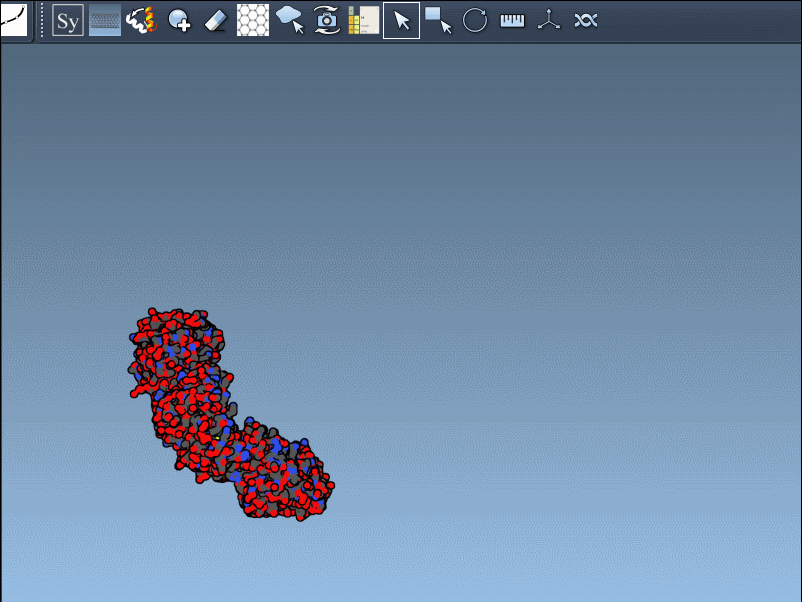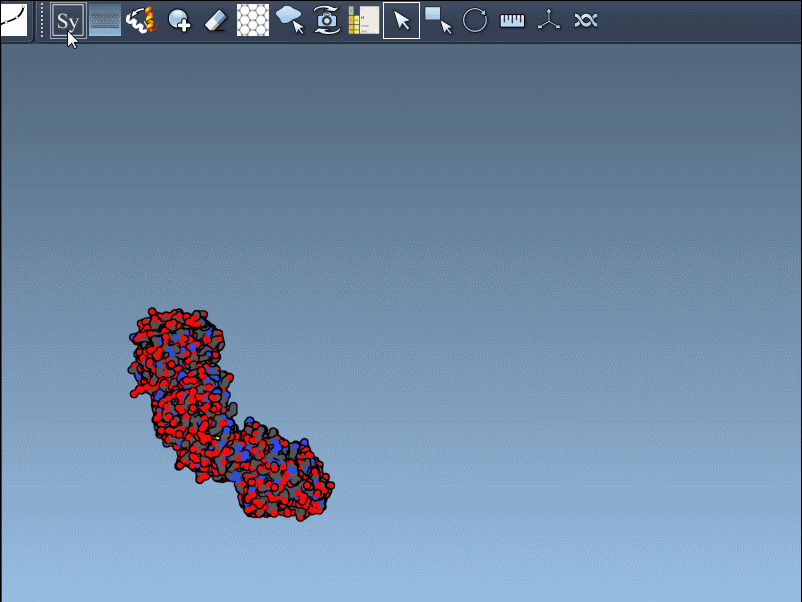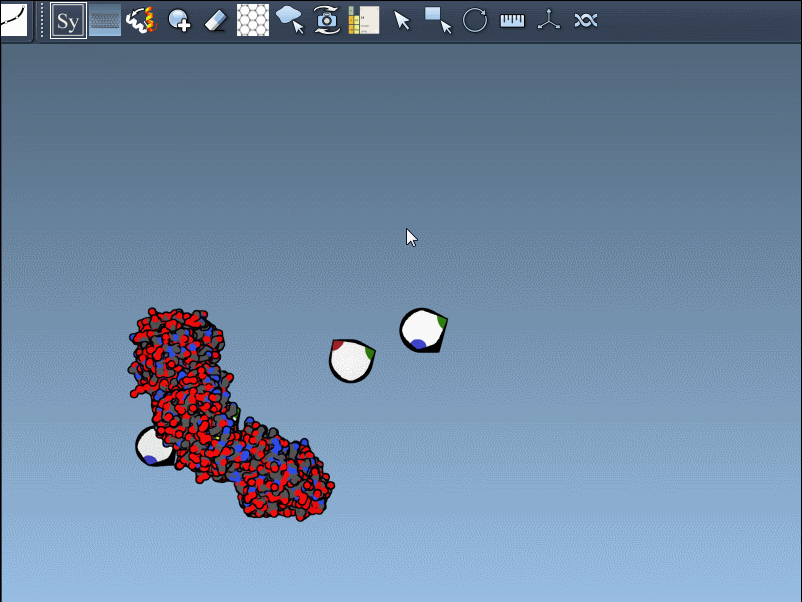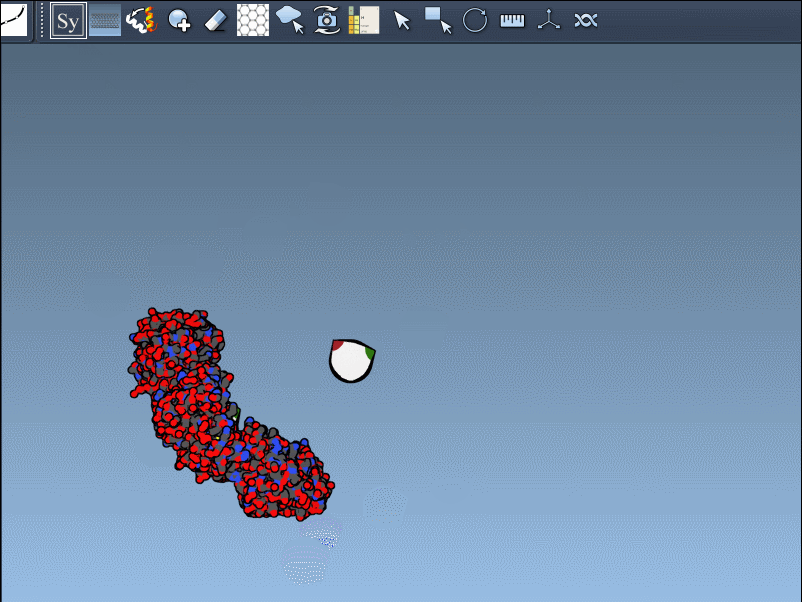
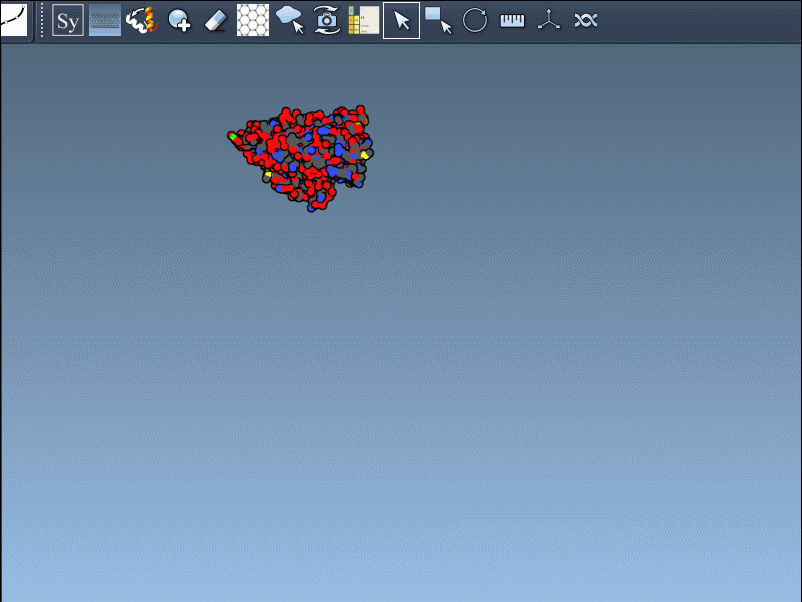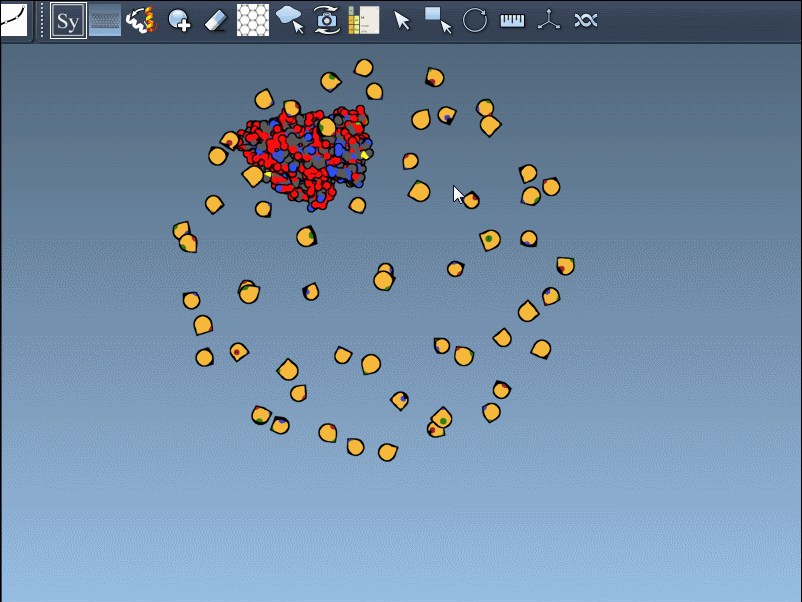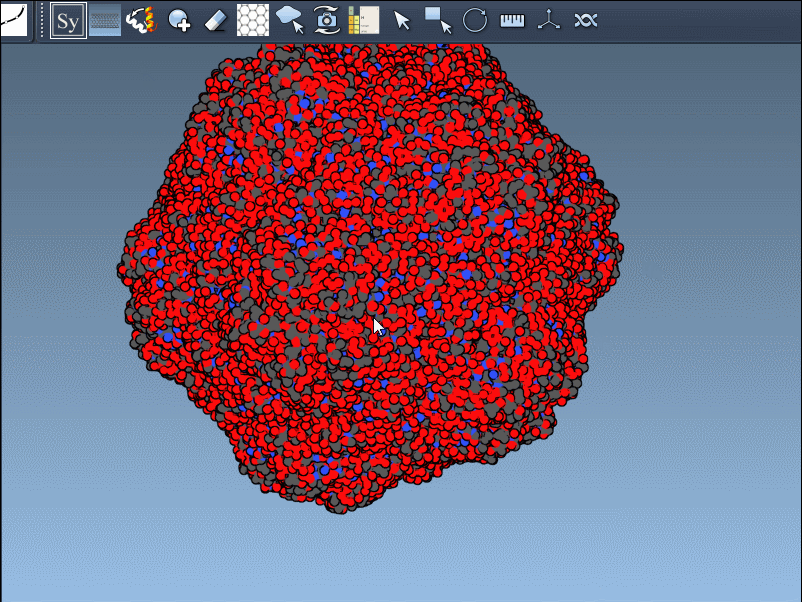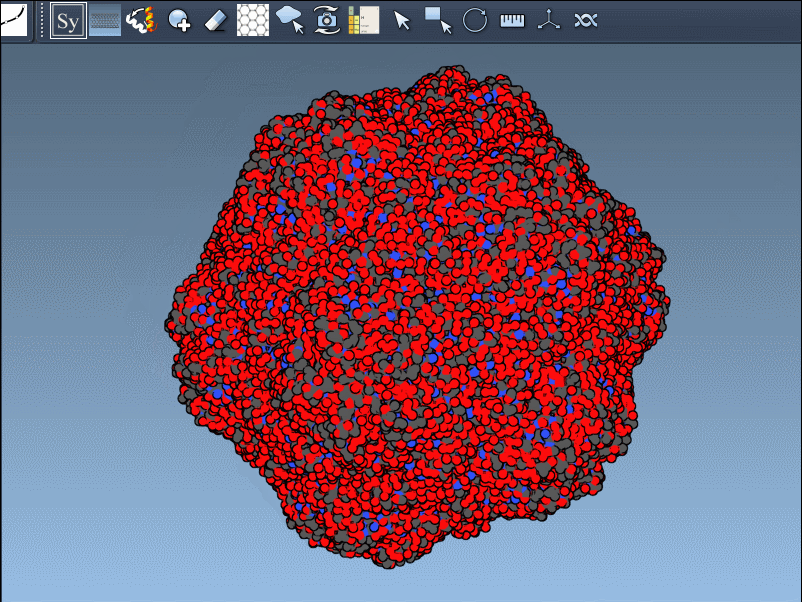
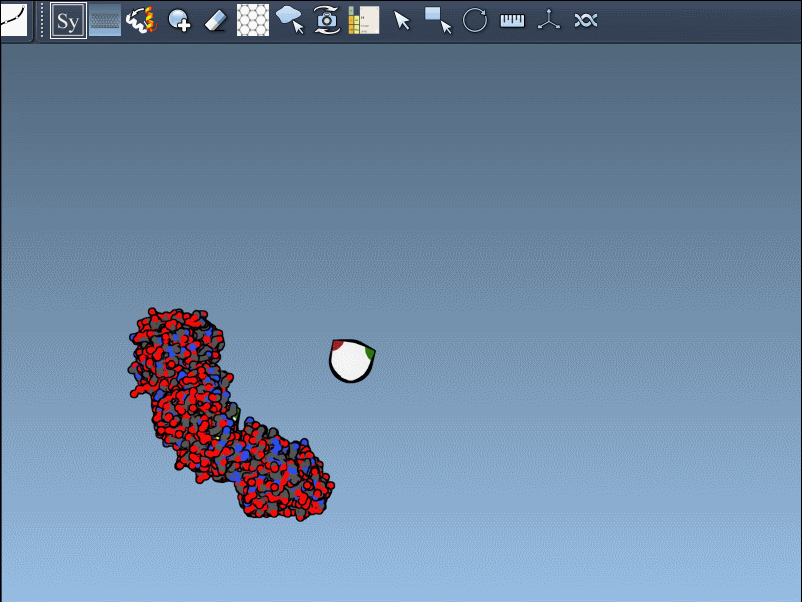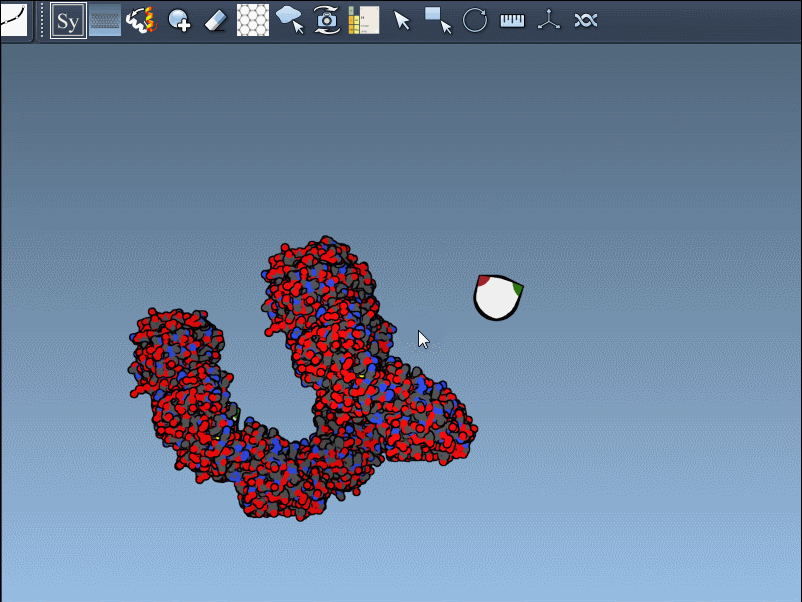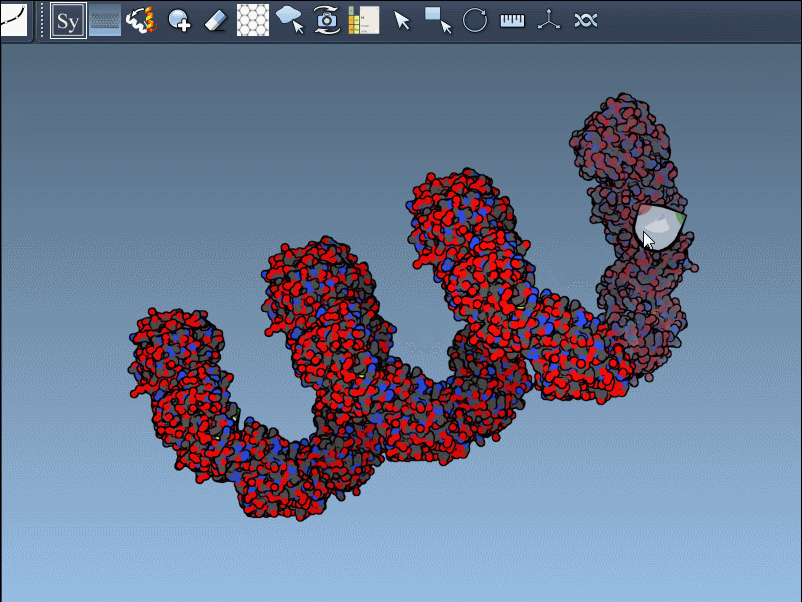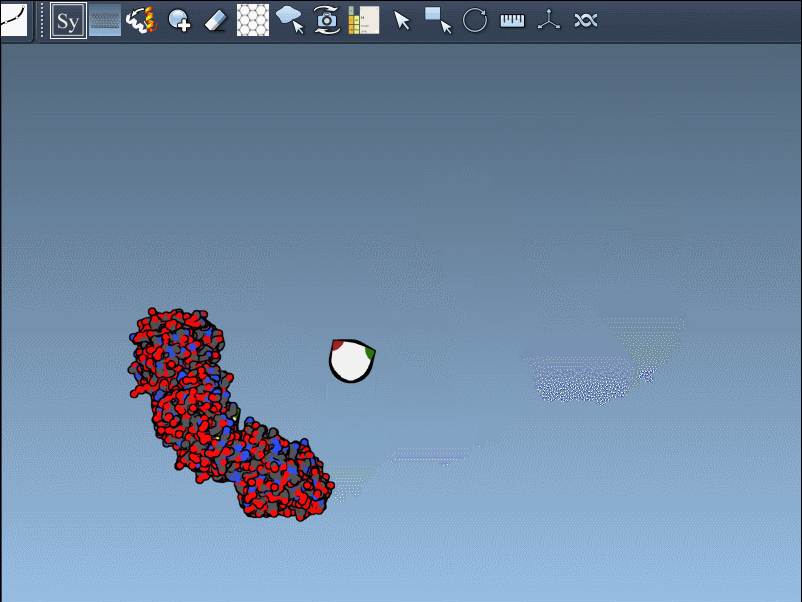
Once you’ve opened a structure in SAMSON and activated the Symmetry Mate Editor, you’ll see control nodes in the viewport. These small interactive widgets represent symmetry operations. Hovering over a node previews the corresponding symmetry mate in real time. This is particularly useful when trying to decide which mates are relevant for your study.
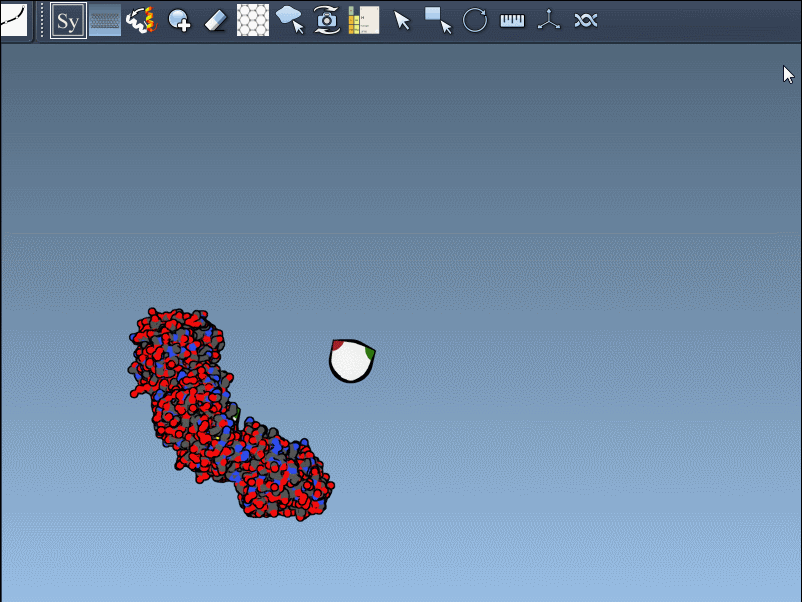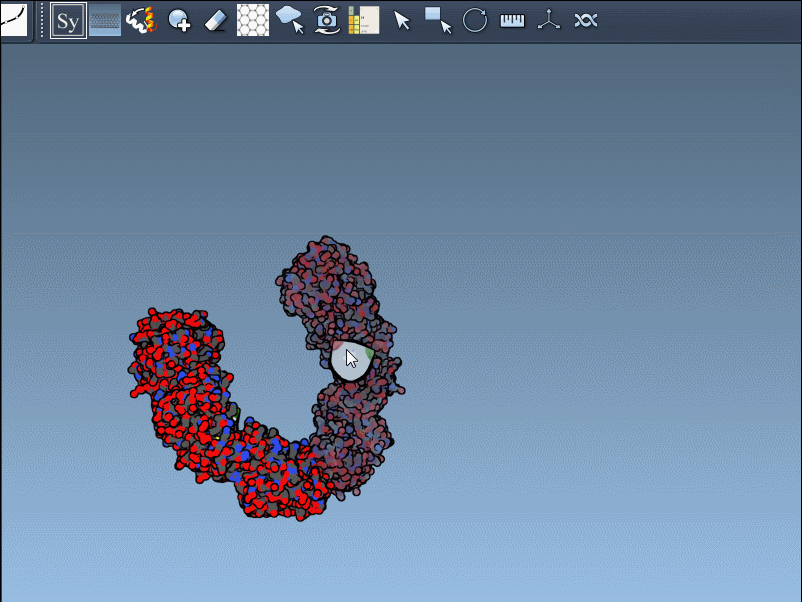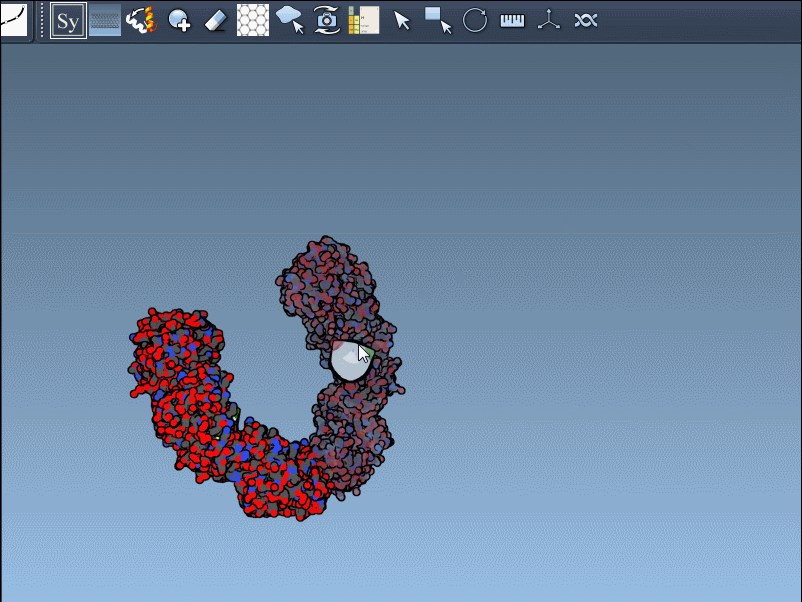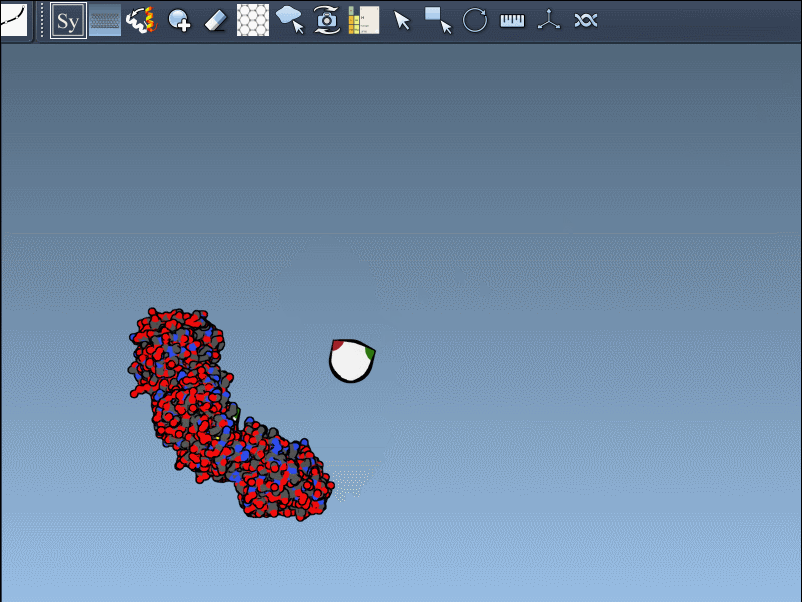
To generate a symmetry mate permanently, simply left-click on the node. It appears immediately in the structure hierarchy—and this makes building more complex assemblies extremely fast.
Scale It Up with Your Mouse
For large symmetries, navigating through dense node networks can be difficult. SAMSON makes it easy to control the number of visible controls with the mouse wheel while holding Ctrl (or Cmd on Mac). This way you can move between a minimal and maximal set of symmetry options with ease.
Preview and Generate All at Once
Need the complete assembly in one go? Hold Ctrl (or Cmd) before hovering and clicking a node, and all related replicas are previewed and generated together. This drastically speeds up the process when working with large or complex symmetric assemblies.
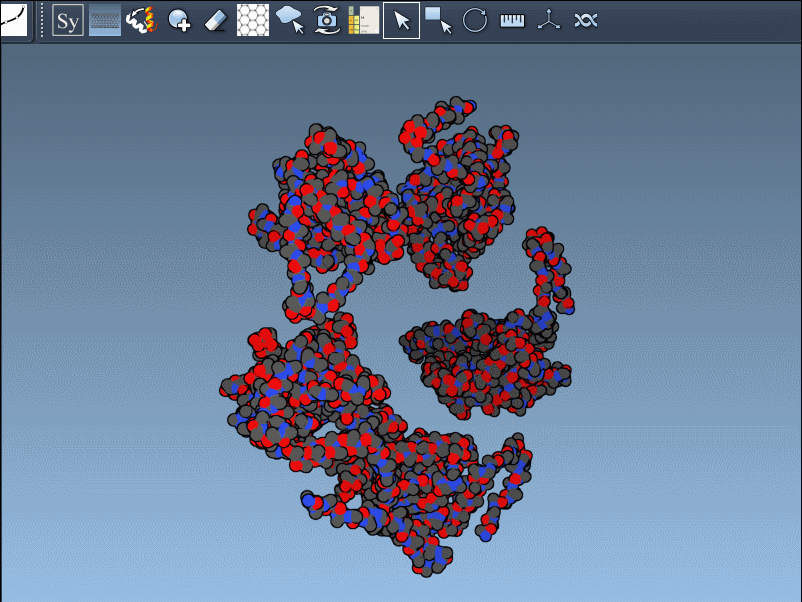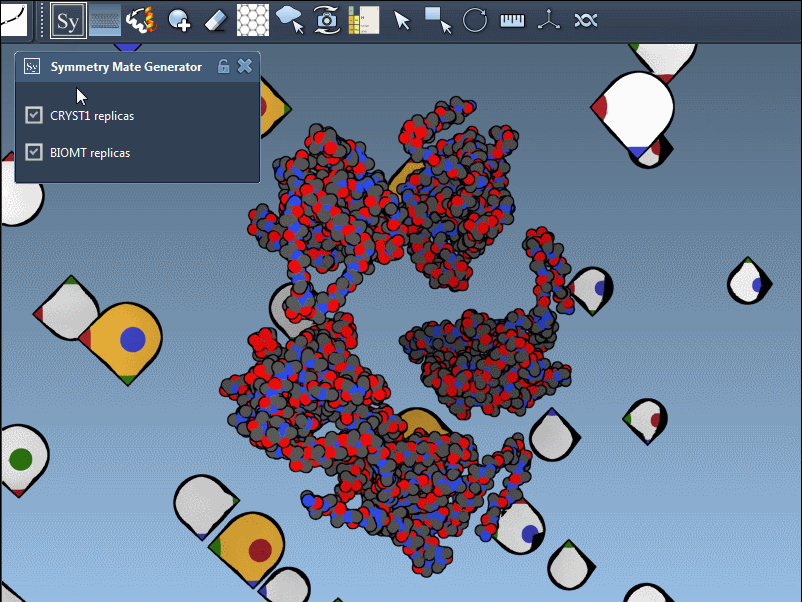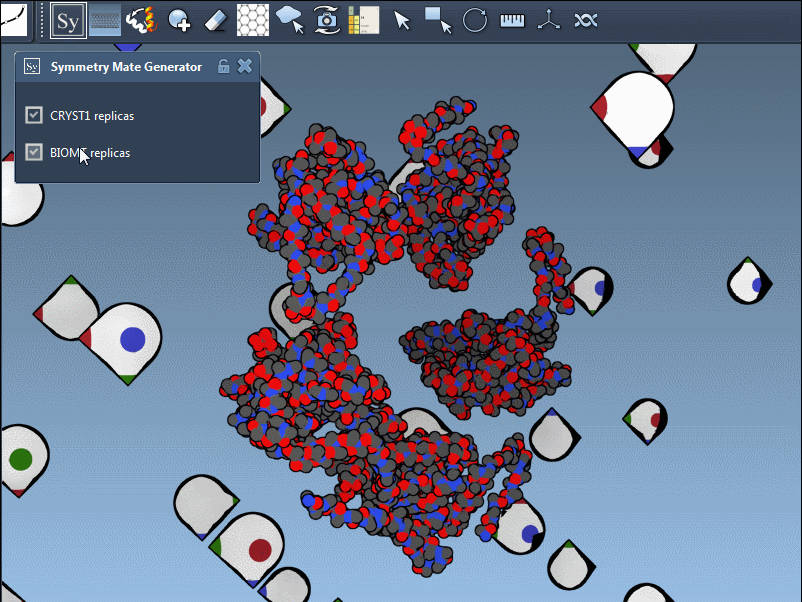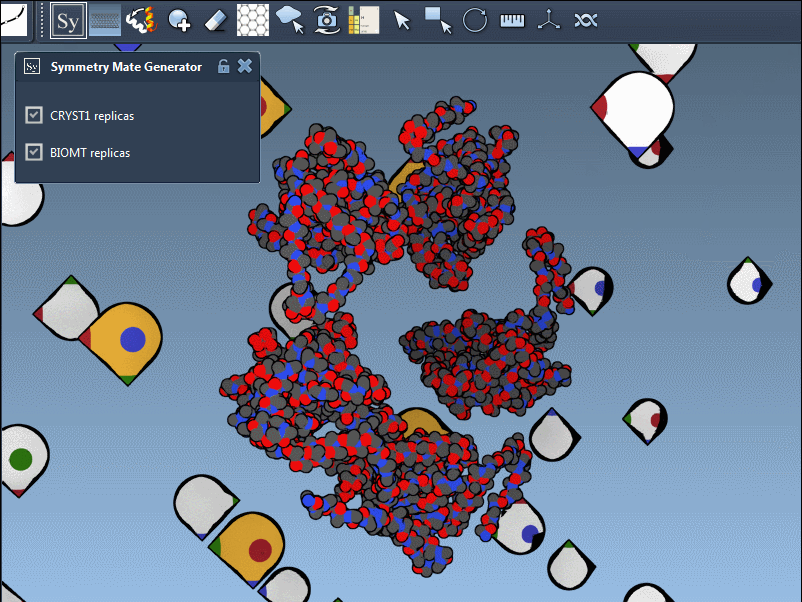
What’s Actually Being Used: CRYST1 vs BIOMT?
When generating symmetry mates, users often forget that PDB files can contain two distinct types of symmetry information:
- CRYST1 records define symmetry based on the crystal lattice.
- BIOMT records are curated annotations indicating the biological assembly.
Using the Symmetry Mate Editor, you can toggle between the two and use them appropriately, depending on whether you’re studying packing or biological function. Here’s an example with different widget colors representing each type:
Undoing Mistakes
And if you generate too many replicas? Simply press Ctrl/Cmd + Z to undo. It’s a subtle but effective way to keep your assembly under control as you experiment.
Conclusion
Interpreting symmetry information in protein structures might seem optional, but often it’s the key to seeing the whole picture. Whether you’re preparing for simulations, studying protein-protein interaction interfaces, or designing symmetric nanostructures, understanding and working with symmetry mates should be part of your everyday modeling workflow.
For step-by-step instructions and other features, see the documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





