When studying complex molecular pathways—like ligand unbinding, conformational transitions, or induced fit mechanisms—molecular modelers often need to extract atomic trajectories along specific paths. But exporting an entire structure is often overkill. What if you’re only interested in a ligand, a backbone, or a binding site residue?
This post demonstrates how to export trajectories of just a selected subset of atoms using the Export Along Paths extension in SAMSON. This improves workflow efficiency and makes follow-up analysis (like free energy calculations) more manageable.
When exporting everything is too much
Imagine you’ve generated a ligand unbinding path using Ligand Path Finder and optimized it using the Parallel Nudged Elastic Band method. For a free energy calculation, you don’t need the full protein trajectory—just the ligand’s.
Luckily, SAMSON’s Export Along Paths tool allows you to export the movement of just the atoms you care about.
Step-by-step: Export selected atoms along a path
Follow this process to stay focused on what matters (your atoms of interest):
- Open the Export Along Paths app from Home > Apps > All > Export Along Paths.
- Under the Advanced panel, select your subset. For example, choose only the ligand—
TDG—from the Document view. - Click Add to register the selected group as a model to export.
- Rename the model if desired (double-click it in the list).
- Ensure one or more relevant paths are selected in the Document view.
- Choose how to export: either a single PDB file containing all frames, or one PDB file per frame.
- Finally, click Export atoms along paths to PDB files and select output location and file prefix.

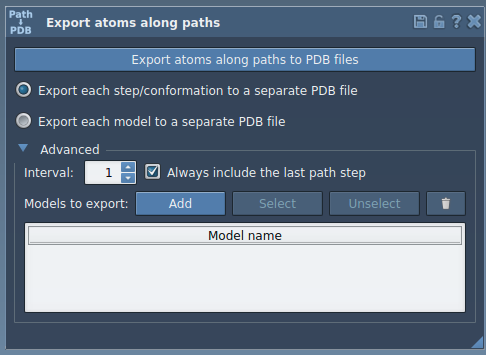
You can define several models (e.g., ligand and nearby residues), include/exclude them dynamically, and toggle them quickly using the interface. This keeps exports clean and tailored to specific modeling tasks.
Why it matters
Exporting only a subset of atoms makes it easier to:
- Prepare input files for umbrella sampling or reaction coordinate studies
- Visualize key conformational changes without background clutter
- Reduce storage and processing time
- Analyze dynamics of regions of functional interest
If you often go from modeling to simulation or back, fine-tuned export control isn’t just a nice-to-have — it’s a necessity.
More helpful tips
You can create and name multiple models for export and return to them later. Use the Select/Unselect commands to highlight them in the scene or use the reset tool (![]() ) to redefine the selection completely.
) to redefine the selection completely.
To learn more about path-based exports in SAMSON, head over to the official documentation:
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





