For molecular modelers working with synthetic or biological polymers, flexibility in defining and reusing molecular building blocks is critical. When creating complex polymers—especially those with repeating units or custom architectures—managing monomer sequences efficiently can save time and reduce errors.
This post focuses on how the Polymer Builder app in SAMSON helps you streamline custom polymer modeling using monomer sequences. If you’re tired of manually assembling long chains or dealing with inflexible templates, this is worth checking out.
Why Use Monomer Sequences?
Monomer sequences are especially useful when designing block copolymers, repeating units in biopolymers, or alternating sequence motifs. Rather than recreating the architecture each time, sequences let you define, visualize, validate, and reuse complex combinations of monomers with full control over bond types and repeat counts.
How It Works
Once you’ve registered individual monomers (i.e., structural fragments like phenyl rings, alkyl chains, peptides, etc.), you can register a sequence by clicking Add new sequence and entering the formula using monomer IDs—like ABBA. Here’s where it gets interesting: bond types can be specified inline with notation like = for double bonds and # for triple bonds:
A=Bmeans A is connected to B with a double bond.A#Bmeans A is connected to B with a triple bond.
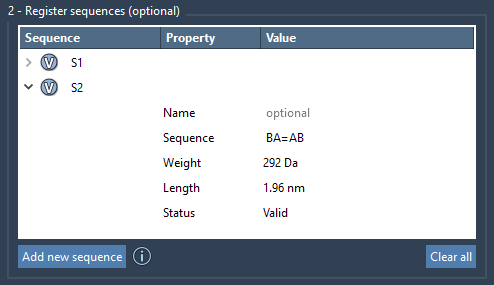
Each registered sequence is auto-labeled (e.g., S1, S2) and can be reused in longer chains. You can also assign descriptive names to group monomers in the polymer’s structure tree, which is useful when tracking sub-blocks of complex polymers.
Error-Checking and Visualization
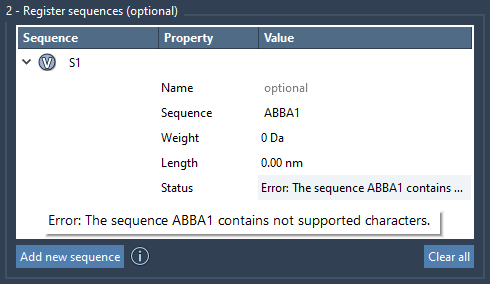
One of the most helpful features for modelers is automatic validation. If there’s an invalid element in your sequence, such as a nonexistent monomer ID or a bonding incompatibility, SAMSON flags it clearly with a status message. For approved sequences, a Valid label appears—and you can even preview what atoms are involved by clicking the V button next to each entry.
Combining and Scaling Sequence Units
Build larger polymers by combining multiple sequences and scaling them in the generation step. For instance:
|
1 |
S1 + 2*S2 + AB=BA + A#B |
This means: one unit of sequence S1, two copies of sequence S2, plus a fragment formed by joining AB and BA via a double bond, followed by a fragment where A and B are joined via a triple bond. This offers you full flexibility over the linear order, repetition, and connectivity of your building blocks.
Approximate molecular weights and polymer lengths update automatically, giving you insight into size and structural features before building the model.
Use Sequences as Building Blocks
Once generated, a polymer can itself become a building block in a higher-level design. Register it as a monomer and define new sequences, enabling efficient top-down design of multiblock or hierarchical polymers.
Conclusion
If you’re managing multi-step polymer builds or working with frequently used structural patterns, using registered sequences inside the Polymer Builder saves time and ensures consistency across simulations or designs. This approach combines clarity with reusability, and it integrates smoothly with other features in SAMSON, including structure minimization, hydrogen adjustment, and export tools.
To learn more, see the full documentation here: https://documentation.samson-connect.net/tutorials/polymer-builder/polymer-builder/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





