Designing analogues and exploring chemical space are central tasks in molecular modeling and drug discovery. However, generating chemical libraries using conventional cheminformatics workflows can often be time-consuming and require custom scripting or external software tools.
The Replace fragments feature of the SMILES Manager extension for SAMSON provides a solution to this pain point. It offers a fast and visual way to build large collections of molecules by simply swapping fragments within a given scaffold—no scripting required.
What does fragment replacement do?
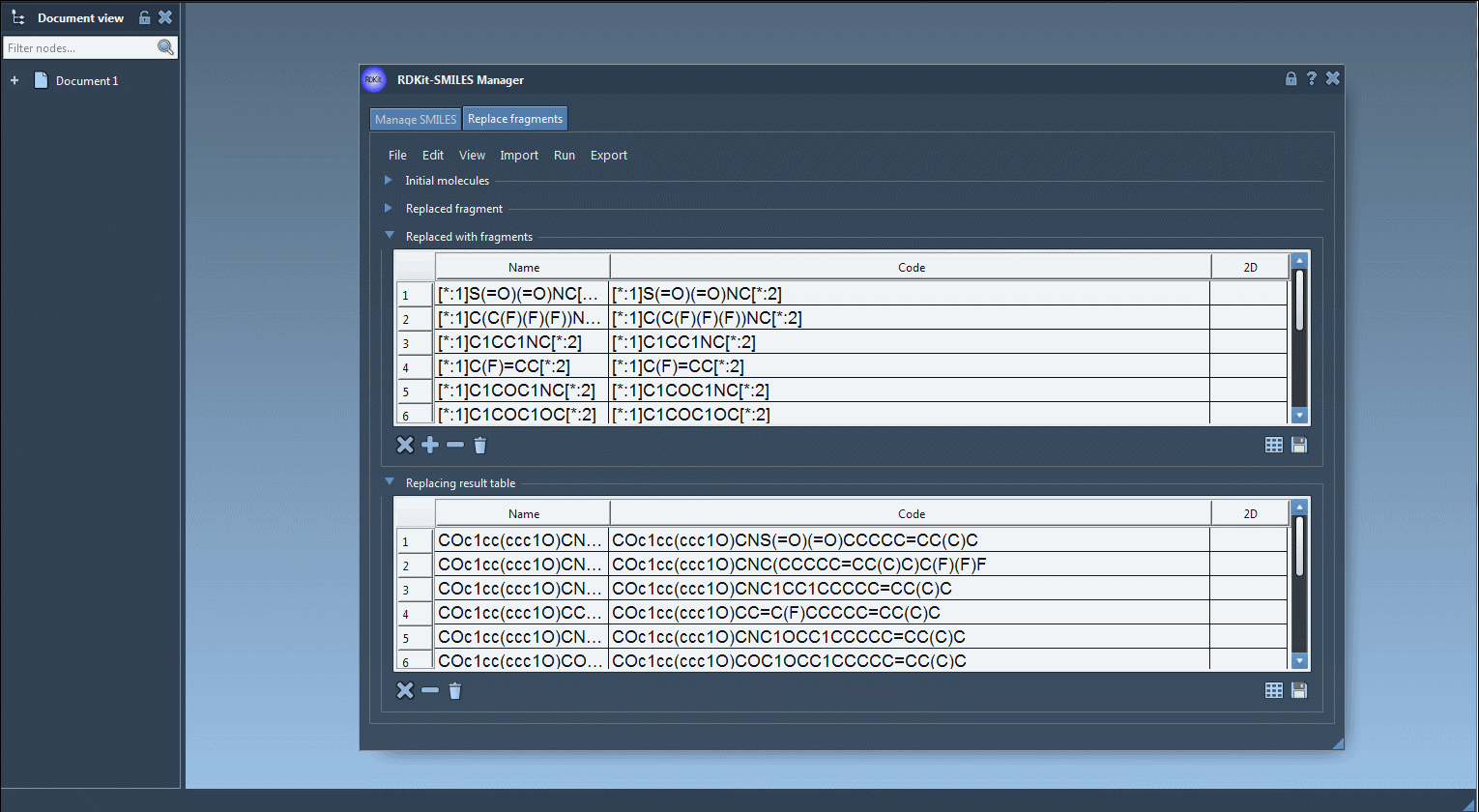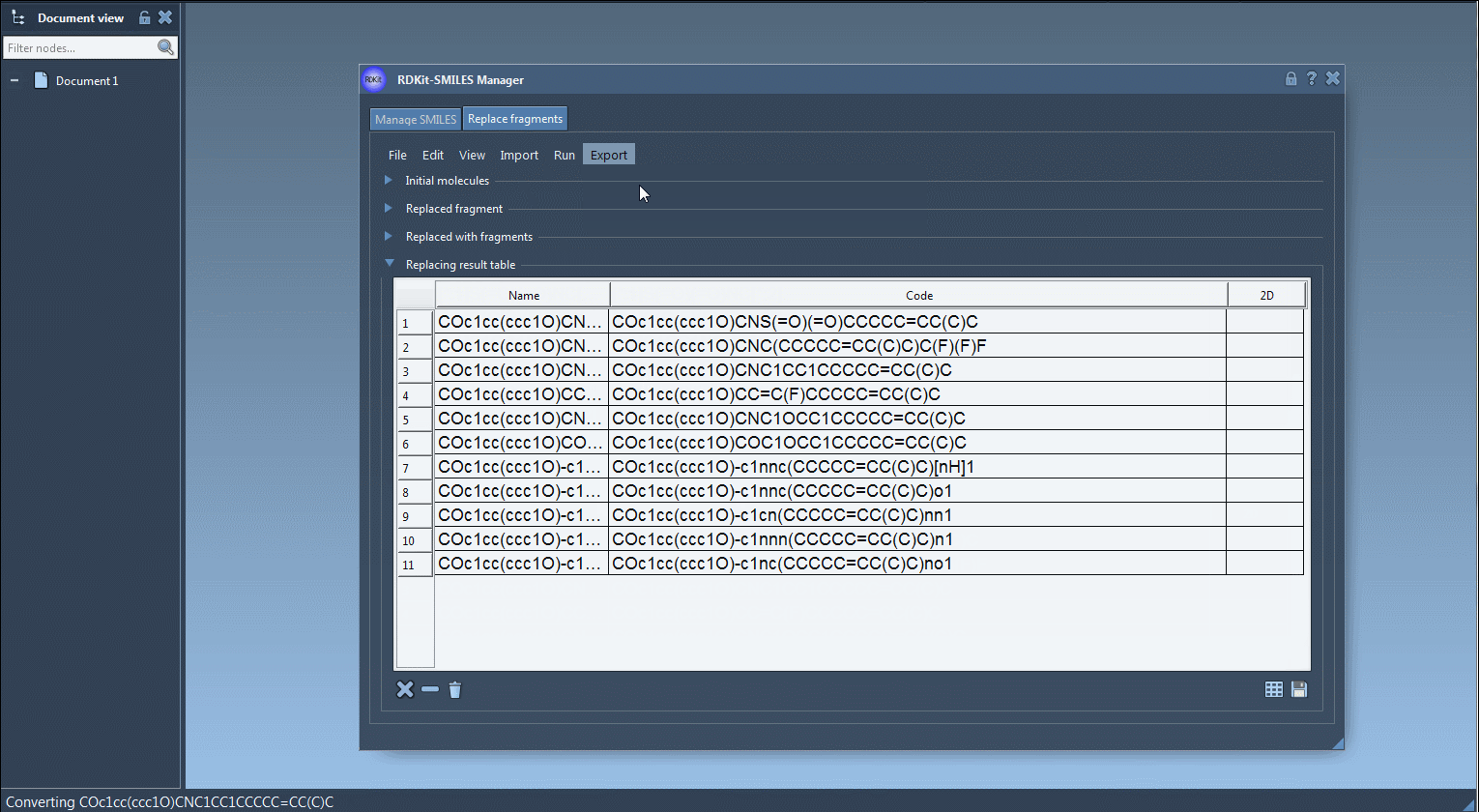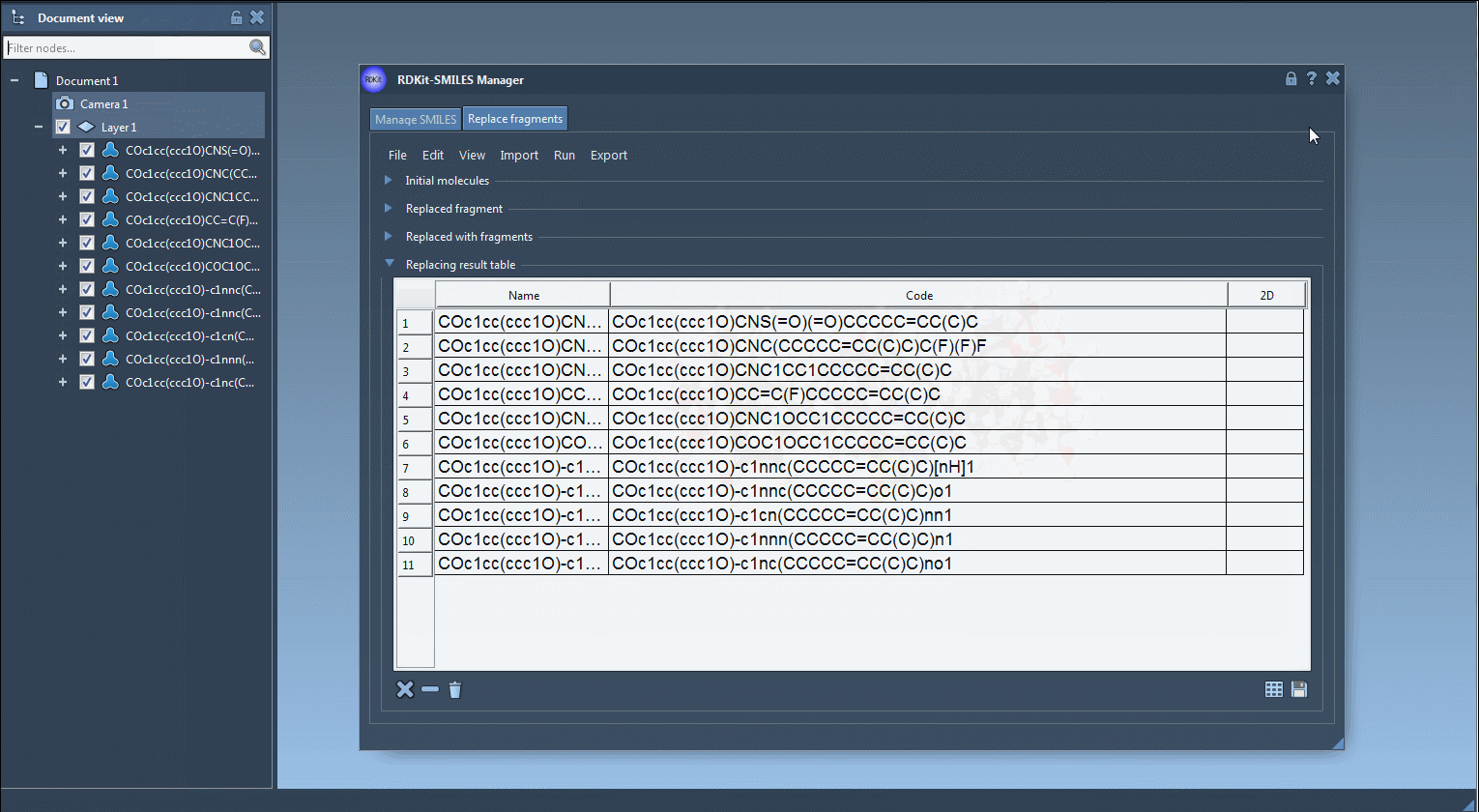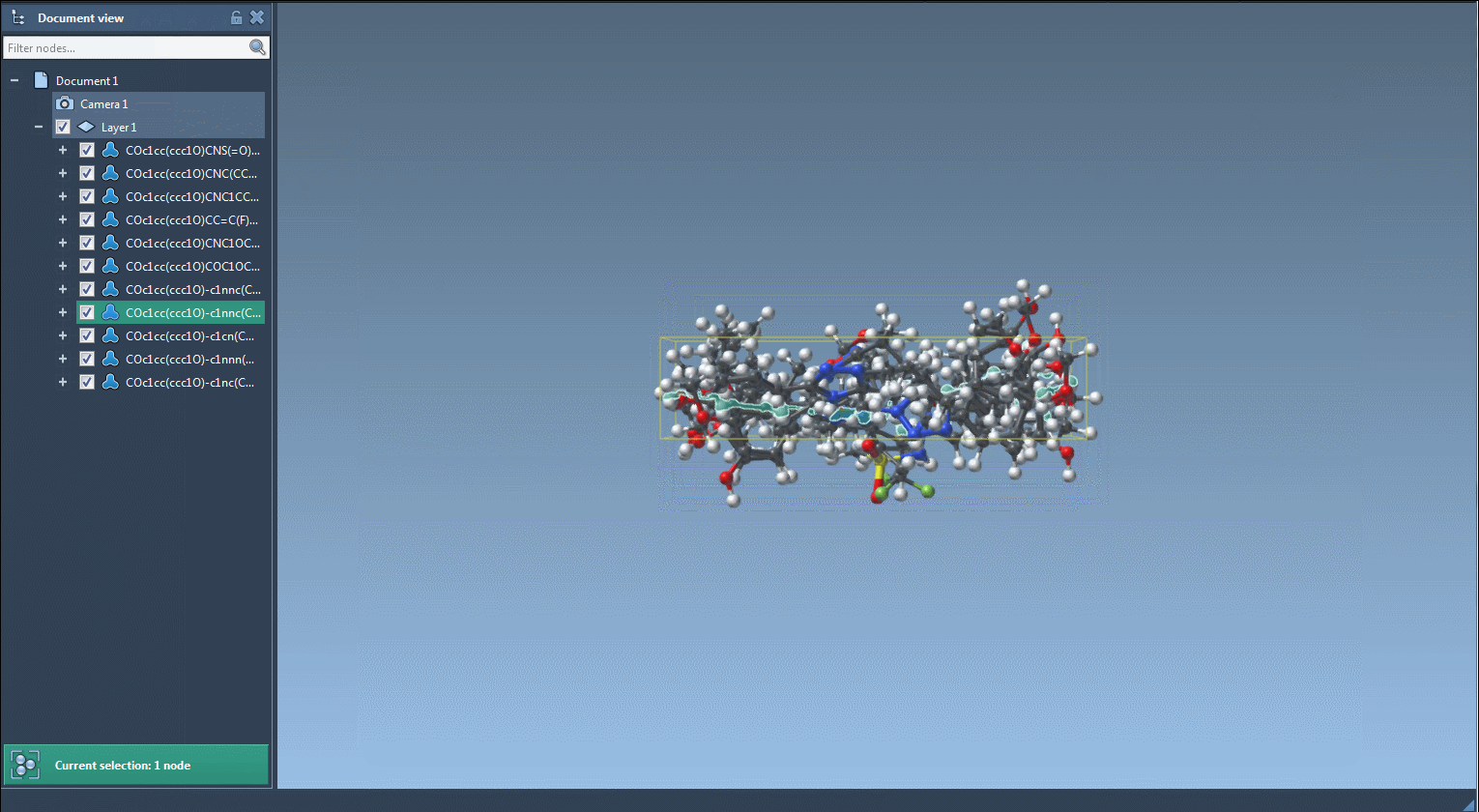
This feature enables you to specify a fragment of a molecule that should be replaced (e.g., a linking group) and define a list of substitute fragments. The SMILES Manager then creates new molecules by replacing the target fragment in each starting molecule with each substitution fragment. The result is a diverse set of analogues that are immediately available for further modeling or export.
This approach is particularly useful for medicinal chemistry applications such as:
- Exploring isosteric replacements
- Optimizing ADME properties by modifying linkers
- Generating combinatorial libraries for screening
How it works in practice
Here’s a concrete example using the SMILES Manager extension:
- Input Molecule: Start with a molecule such as:
COc1cc(CNC(=O)CCCC/C=C/C(C)C)ccc1O - Fragment to Replace: Specify a pattern like:
[:*1]C(=O)NC[:*2]
Note: The[:*1]and[:*2]labels define the fragment attachment points. - Replacement Fragments: Load or define fragments such as:
[:*1]S(=O)(=O)NC[:*2]
[:*1]C(C(F)(F)(F))NC[:*2]
[:*1]C1=NN=C([:*2])N1 - Run Replacement: Click the “Replace fragments” button in the Run menu.
The results will appear in a table format with 2D depictions and names.
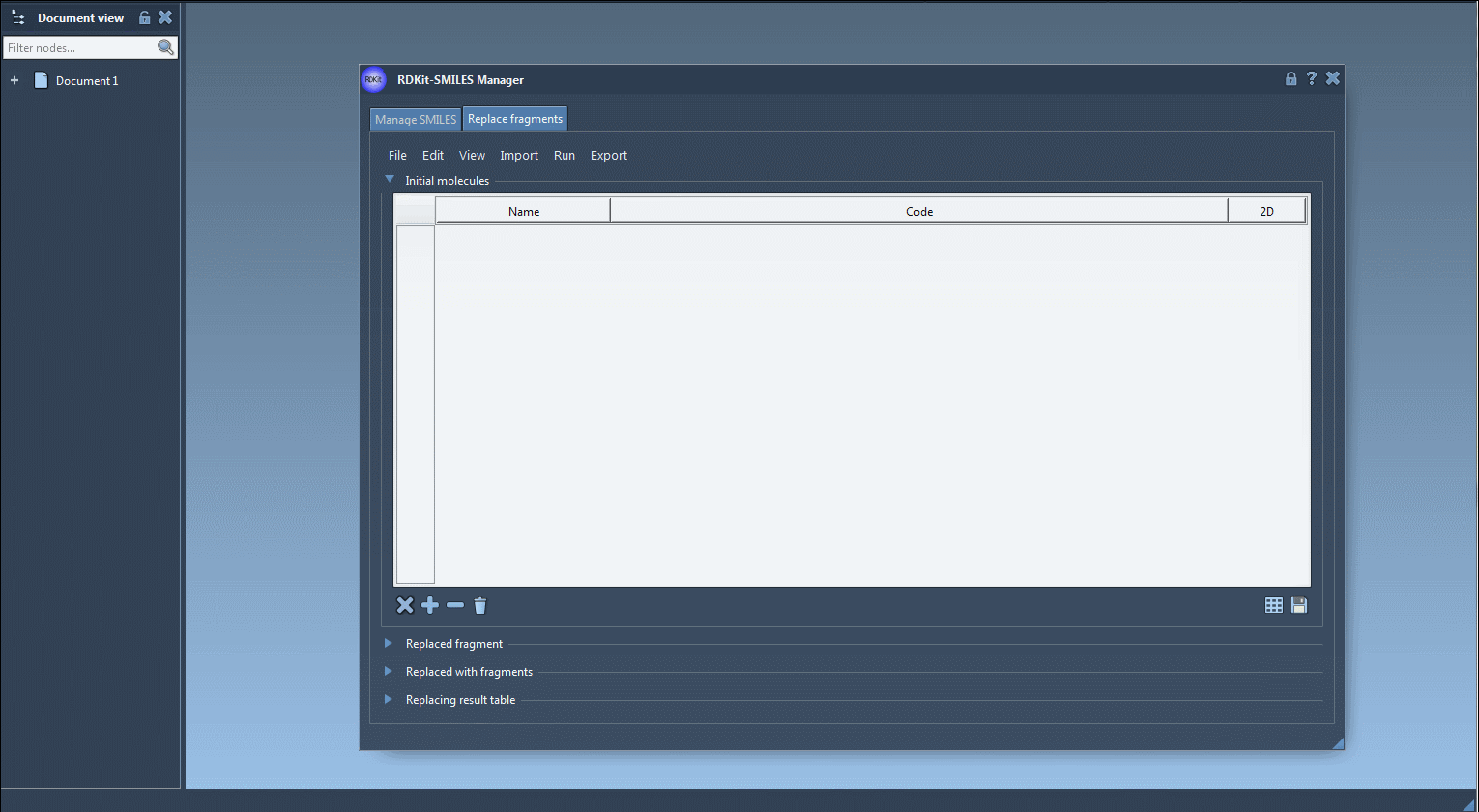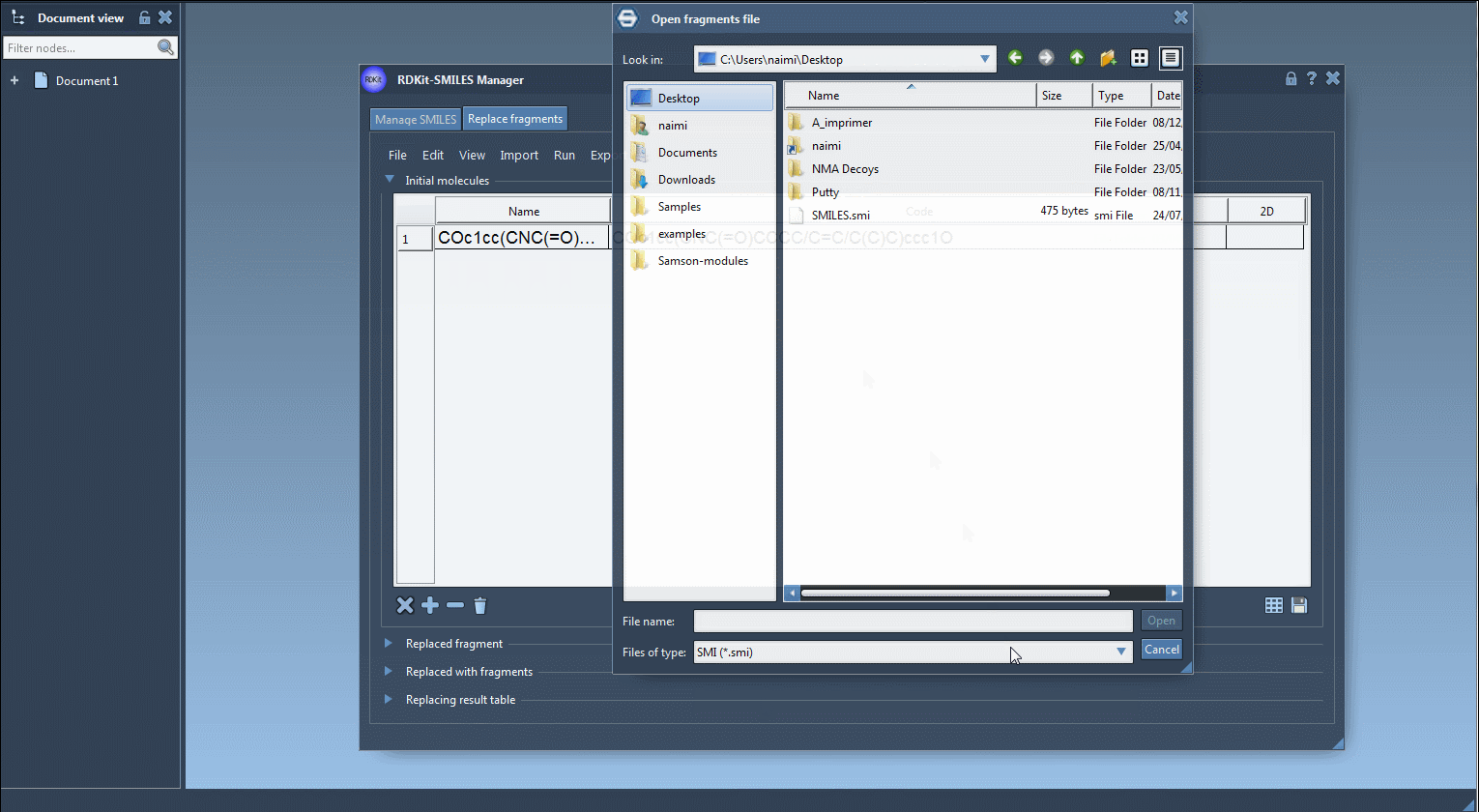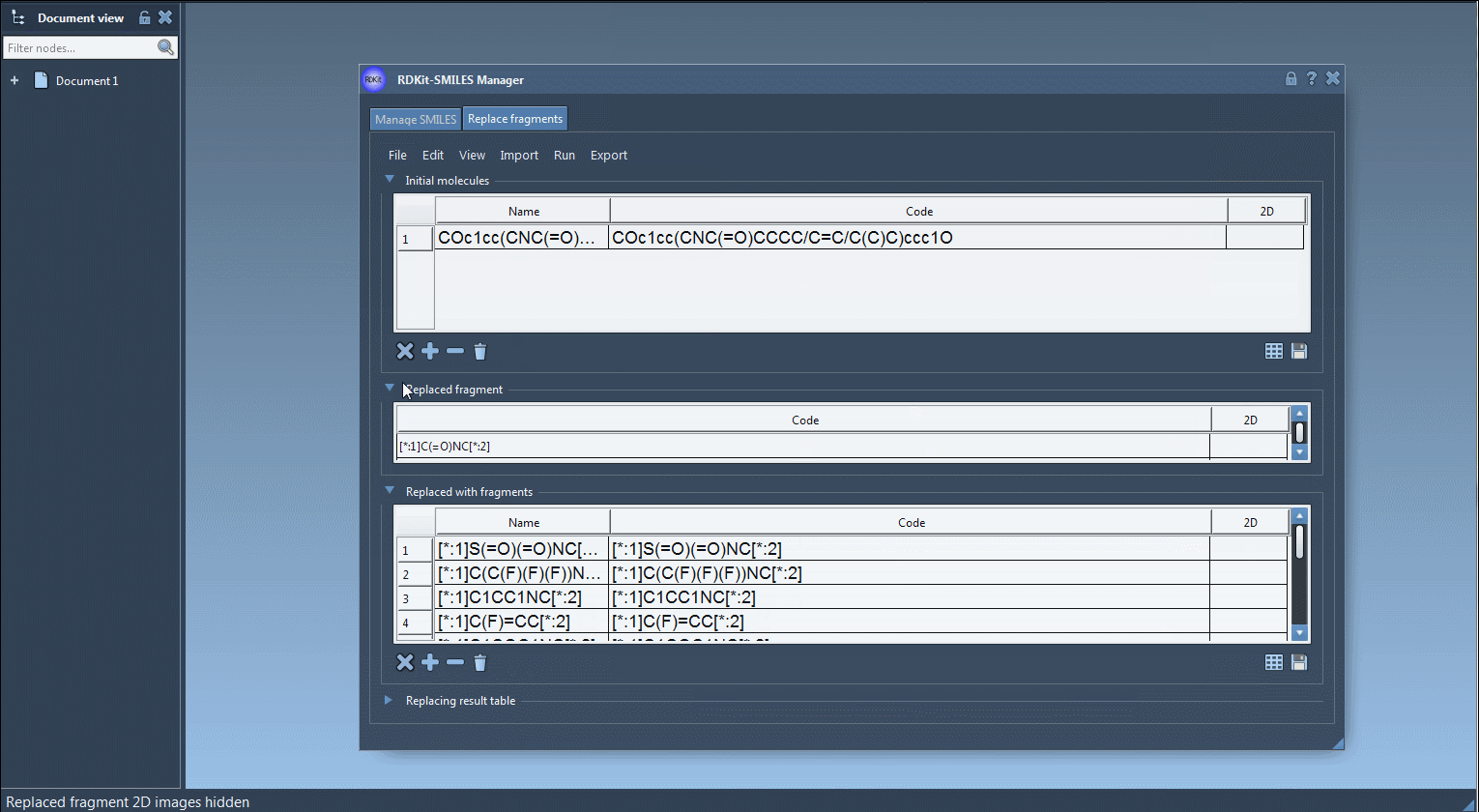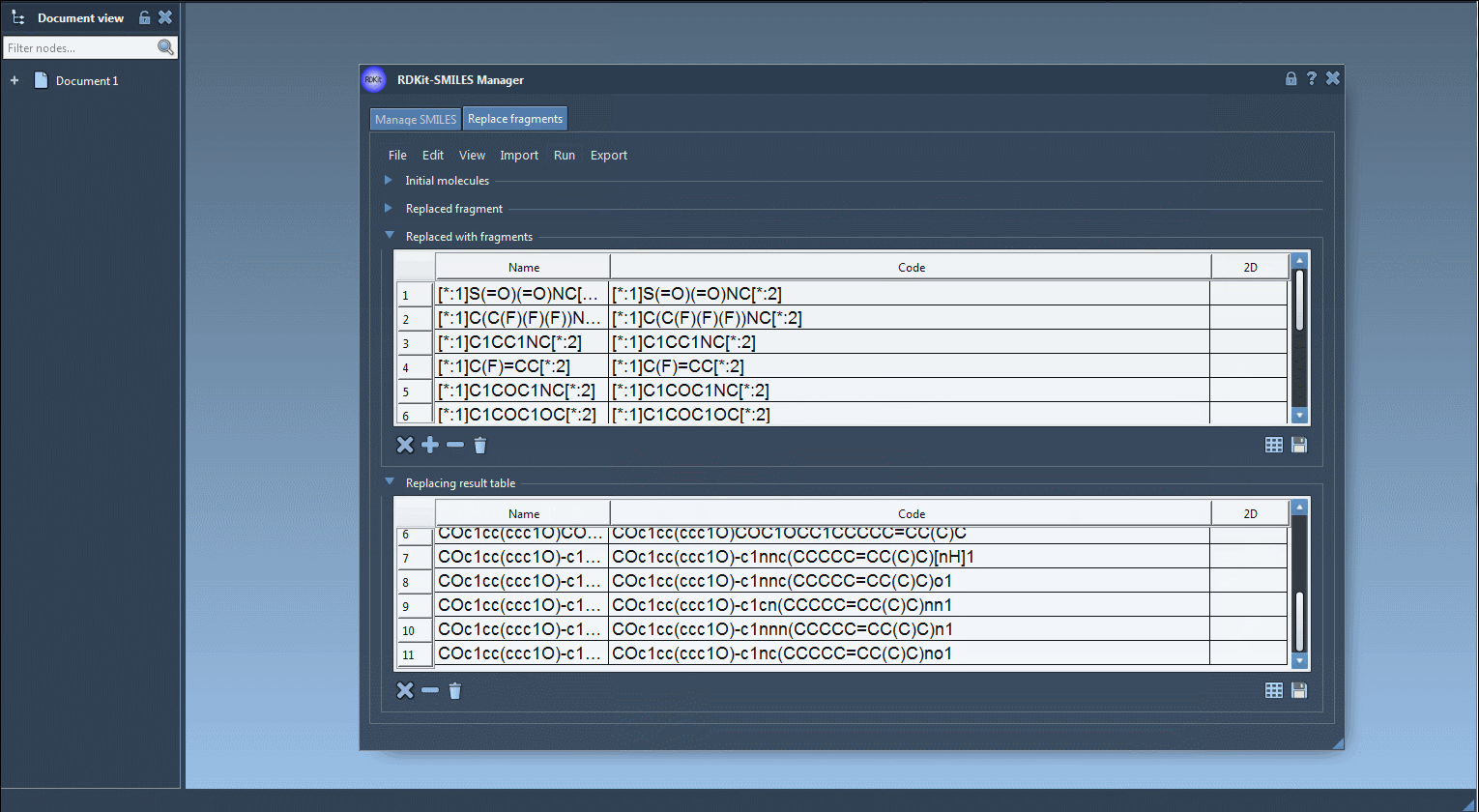
The generated molecules can then be converted into 3D structures using RDKit functions built into the extension. This means you can use the molecules right away in your modeling workflow in SAMSON.
Why this saves you time
Instead of creating analogues one at a time or writing Python scripts to perform SMILES-based replacements, SAMSON’s SMILES Manager lets you control library design visually, while still offering compatibility with text-based input files. It also supports importing molecules or fragments from active SAMSON documents via selections, offering a flexible interface for users who work across both code and structure.
Results can be exported, saved as SVG or PNG grid images, or directly inserted into your SAMSON workspace for visualization, simulation, or additional editing.
Want to try it out?
If you’re designing analogues, evaluating substituent effects, or building SAR libraries, this feature is definitely worth exploring. Check out the full tutorial on the SAMSON documentation portal to see more examples and advanced usage:
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at www.samson-connect.net.





