The ability to track and analyze atom trajectories along molecular pathways is an essential tool in molecular modeling. Whether preparing input files for reaction coordinate studies, performing free energy calculations, or studying ligand binding/unbinding pathways, exporting precise atomic coordinates is often a key challenge. This is where SAMSON’s Export Along Paths extension provides a clear and practical solution.
Why Molecular Modelers Need This
When working with complex molecular systems, modelers frequently need to retrieve atomic coordinates along specific pathways. For example, in free energy profiling or reaction coordinate generation, it’s crucial to track the movements of specific atoms, such as ligands or key residues, at regular intervals.
Manually exporting such data can be extremely time-consuming and prone to errors. With the Export Along Paths app in SAMSON, you can now automate this process with precision and flexibility.
Getting Started: Open, Select, Export
The process of exporting trajectories in SAMSON is user-friendly and efficient. Start by opening the Export Along Paths app from Home > Apps > All > Export Along Paths or using a quick shortcut ( Shift + E ).
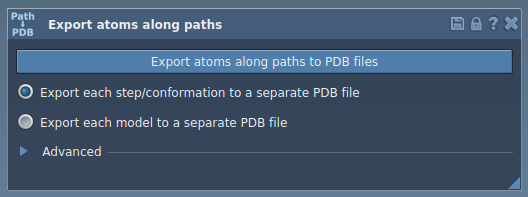
With the app interface open, you can choose to export either:
- All atoms: Export every atom’s trajectory along the pathway in a single or multiple PDB files.
- A specific subset: Focus on selected groups of atoms, such as a ligand or a defined subset, for targeted analysis.
Step-by-Step Approach to Exporting Atom Trajectories
Option 1: Export All Atoms
- Select one or more paths from the Document view.
- Choose an export format: single PDB file containing all frames, or separate PDB files for each frame.
- Click Export atoms along paths to PDB files and select a destination folder and file prefix. Optionally, you can expand the Advanced settings to adjust the export frequency (e.g., exporting every nth frame).
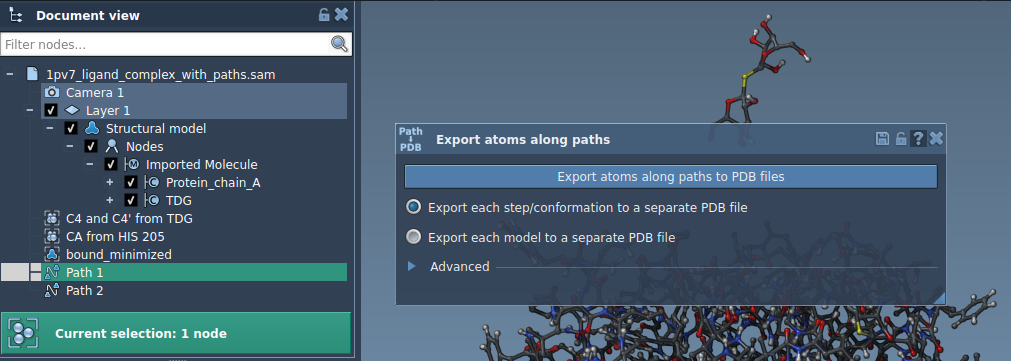
Option 2: Export a Subset of Atoms
- Expand the Advanced panel in the app.
- Select a subset of atoms (e.g., ligand
TDG) using the Document view, and click Add to define the chosen group as a model for export. - Rename the selection, add further groups for export if needed, and finalize your model list.
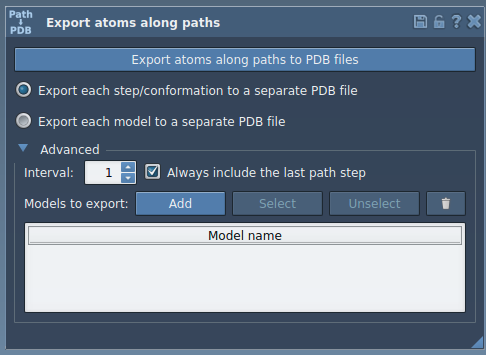

Name your models, select the relevant path(s), and proceed to export. Outputs can again be configured to a single or multiple PDB-file format for downstream use.
Applications in Molecular Science
This feature is particularly impactful for:
- Preparing reaction coordinate files for free energy calculations.
- Exporting ligand exit/entry trajectories to enhance sampling workflows.
- Tracking intermediate states for visualization and analysis.
- Exporting detailed motion trajectories for specific components like active sites, backbones, or ligands.
With the clean, user-friendly interface of the Export Along Paths extension, molecular modelers can spend less time extracting data and more time drawing critical insights from simulations.
Learn more about the full capabilities and applications of this feature by visiting this detailed tutorial.
Note: SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





