For molecular modelers, efficiently exploring the effects of structural modifications on molecular properties can be a challenging task. Whether you’re designing ligands or optimizing bioactivity, pinpointing the optimal molecular analog can feel like searching for a needle in a haystack. That’s where SAMSON’s Positional Analogue Scanning functionality, powered by the SMILES Manager extension, comes in to simplify and expedite your process. Let’s explore a practical walkthrough of how you can use this tool to engineer analogs and analyze their potential.
The Problem: Iterative Molecular Modifications
Testing molecular modifications often involves complex workflows and time-intensive analyses. Manually generating analogs for evaluation can be tedious, particularly when replacing specific patterns within a molecule or attaching functional groups at desired positions. Positional Analogue Scanning (PAS) solves this pain point by pinpointing how these changes affect molecular properties—whether it’s binding affinity, stability, or interactions with a target like a protein.
How It Works: Theory Meet Practice
Begin with a single molecule, either entering its SMILES code directly into the interface or selecting it in your SAMSON project and clicking the Use selection button. As you’re defining your molecule of interest, SAMSON conveniently provides a visual confirmation of your chosen structure, ensuring accuracy.
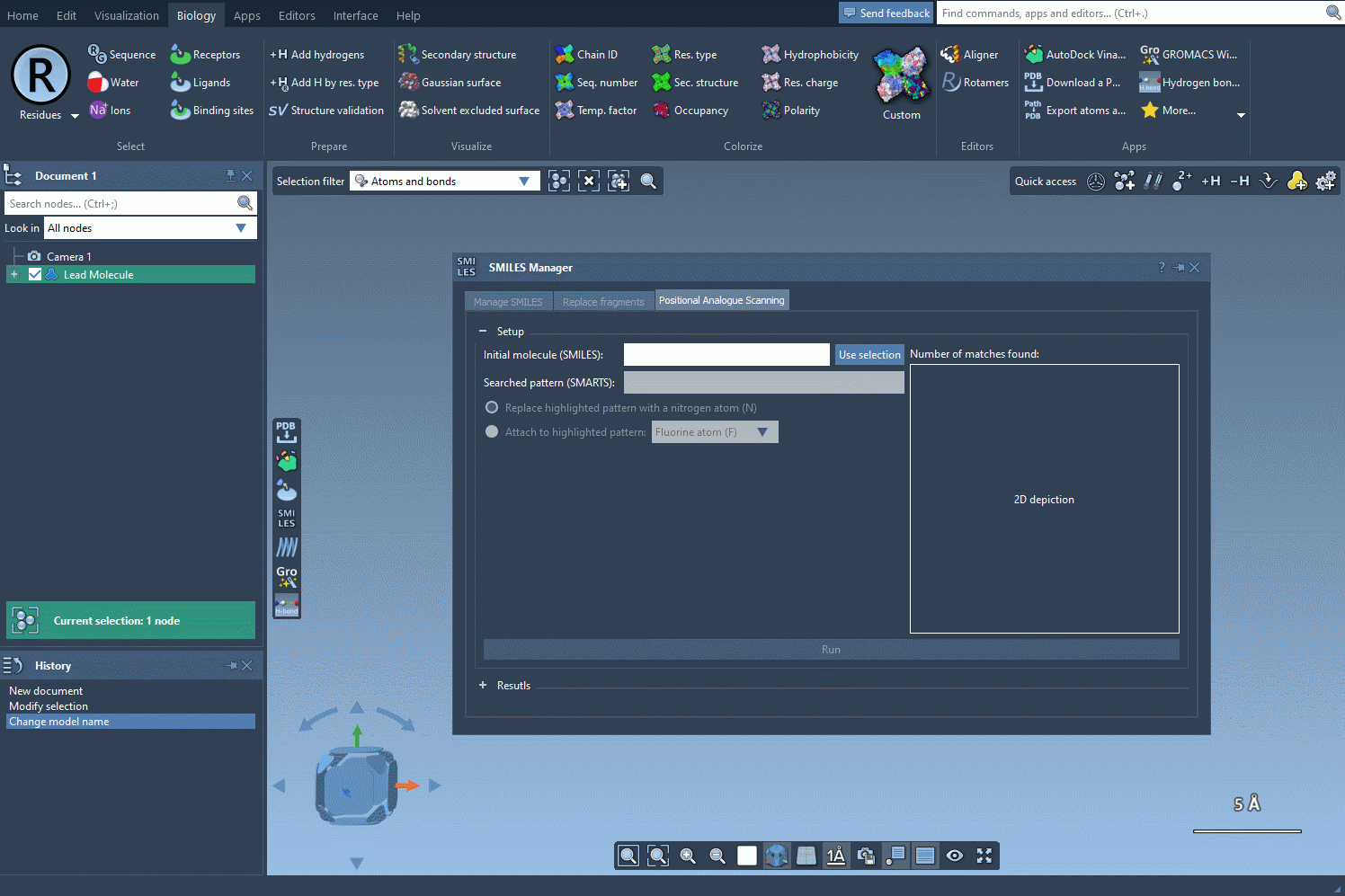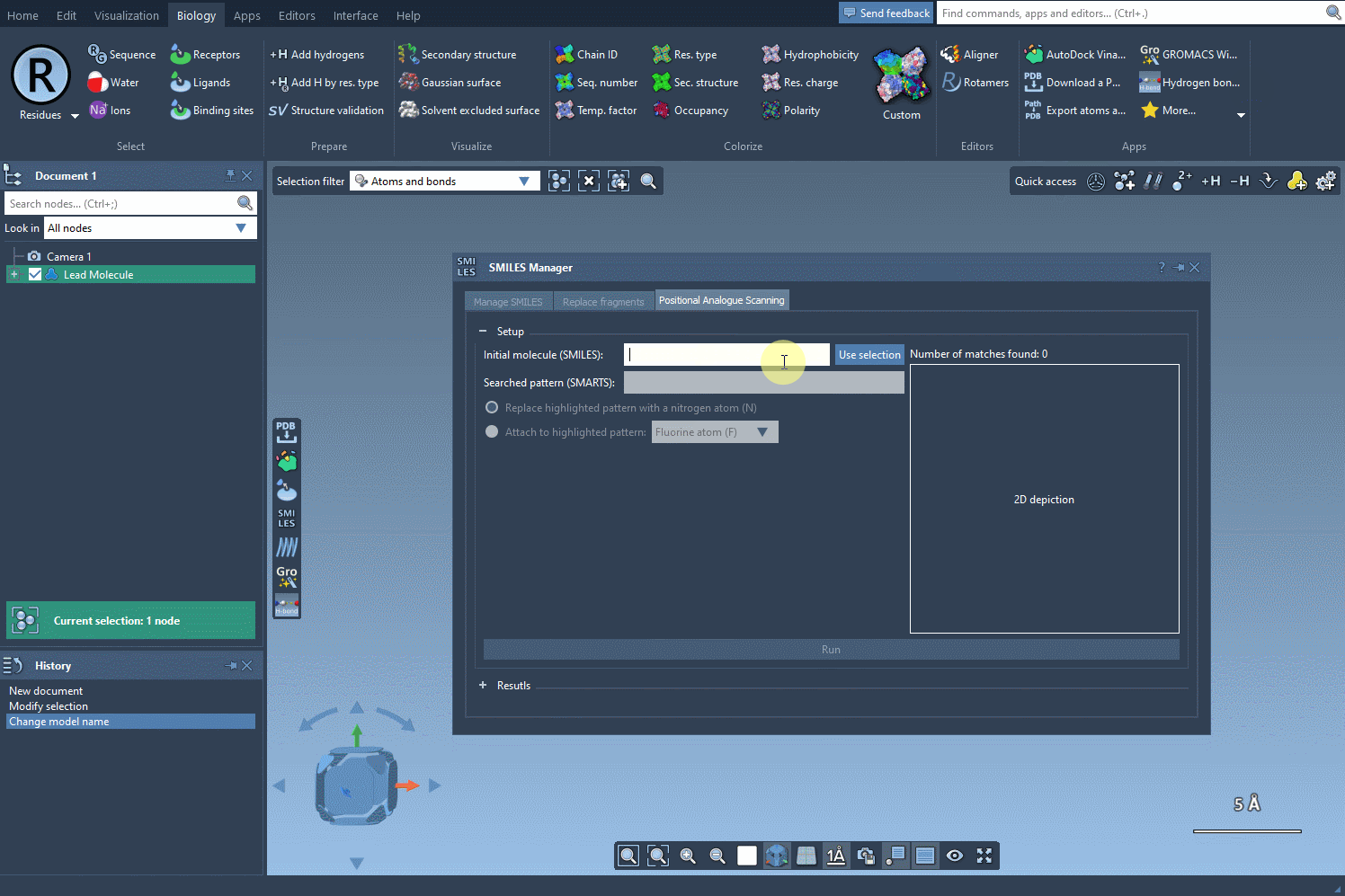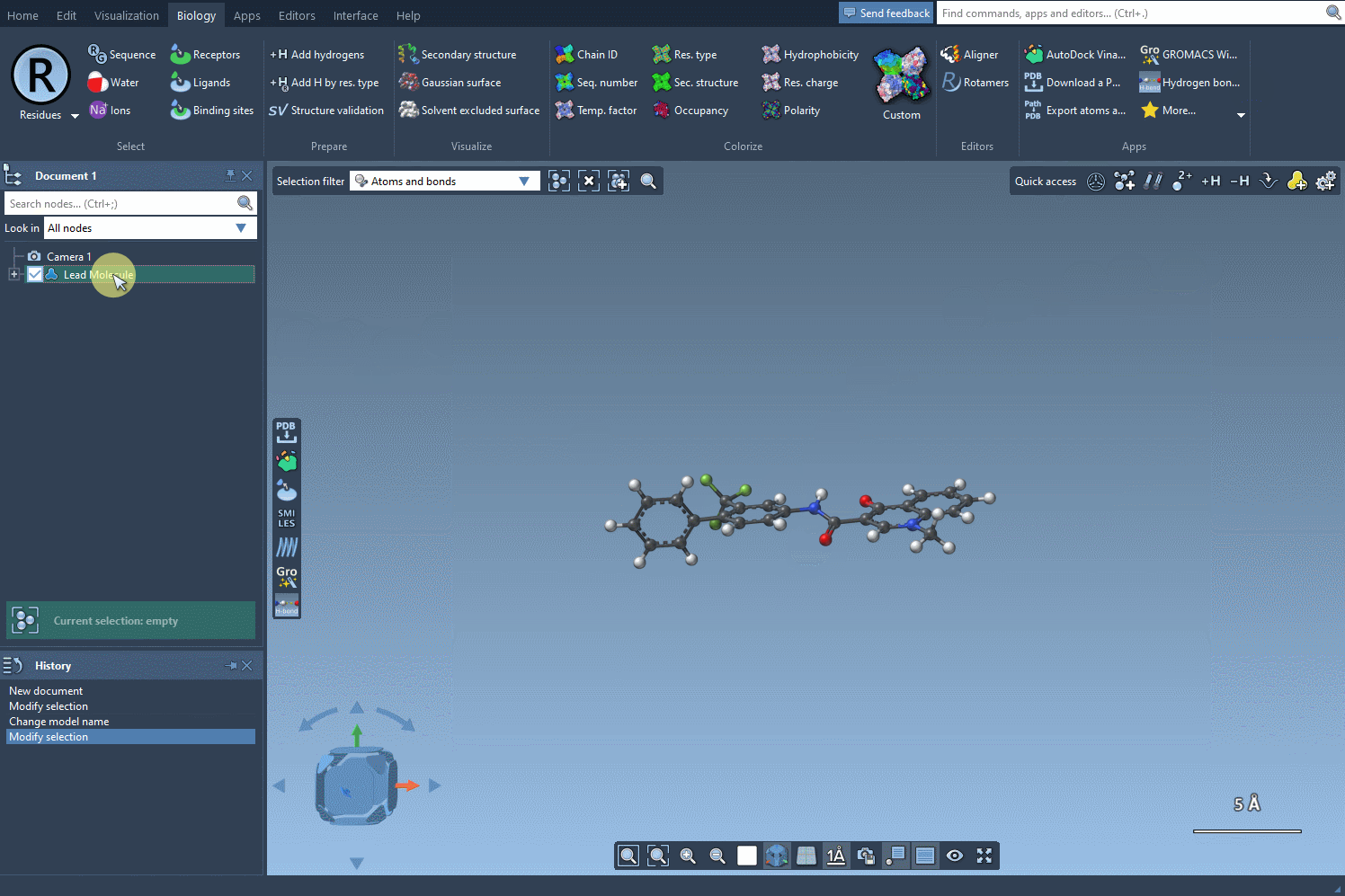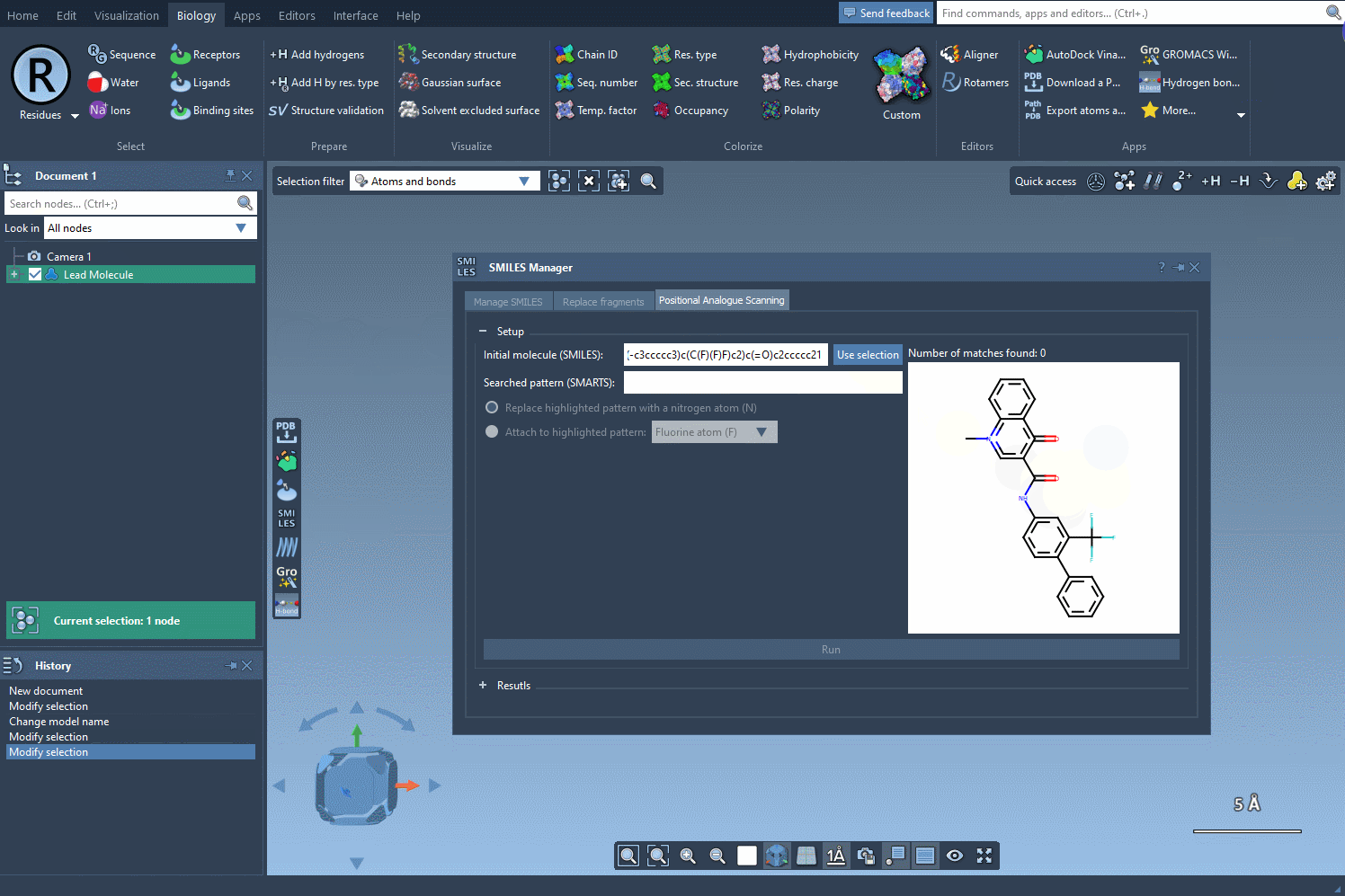
Once the base structure is in place, it’s time to specify the pattern you want to explore. For instance, if targeting aromatic positions, you might use the SMARTS code [cH]. SAMSON’s SMILES Manager instantly highlights these relevant areas in the molecule for you to review:

Pattern Replacement: Immediate Possibilities
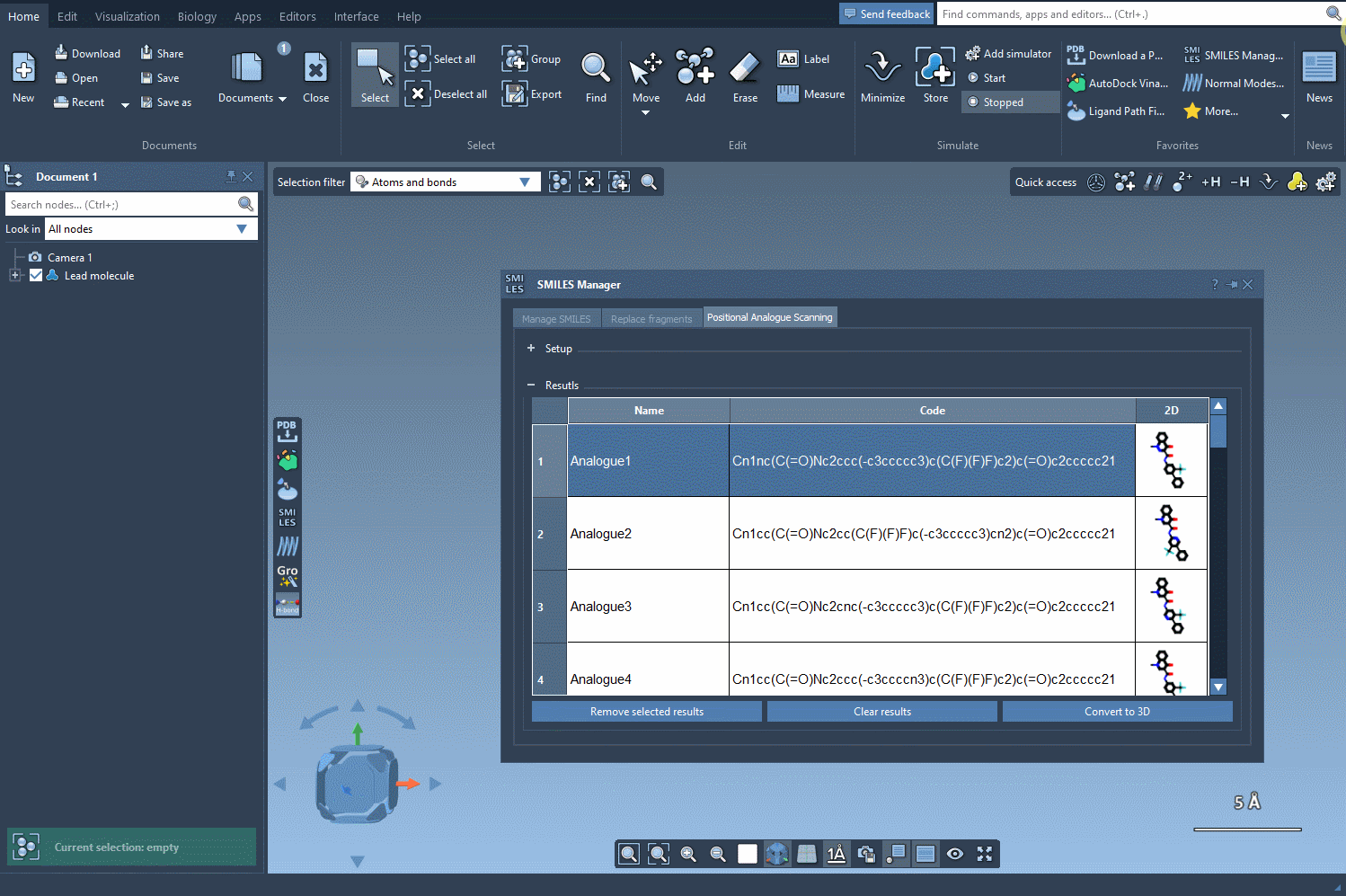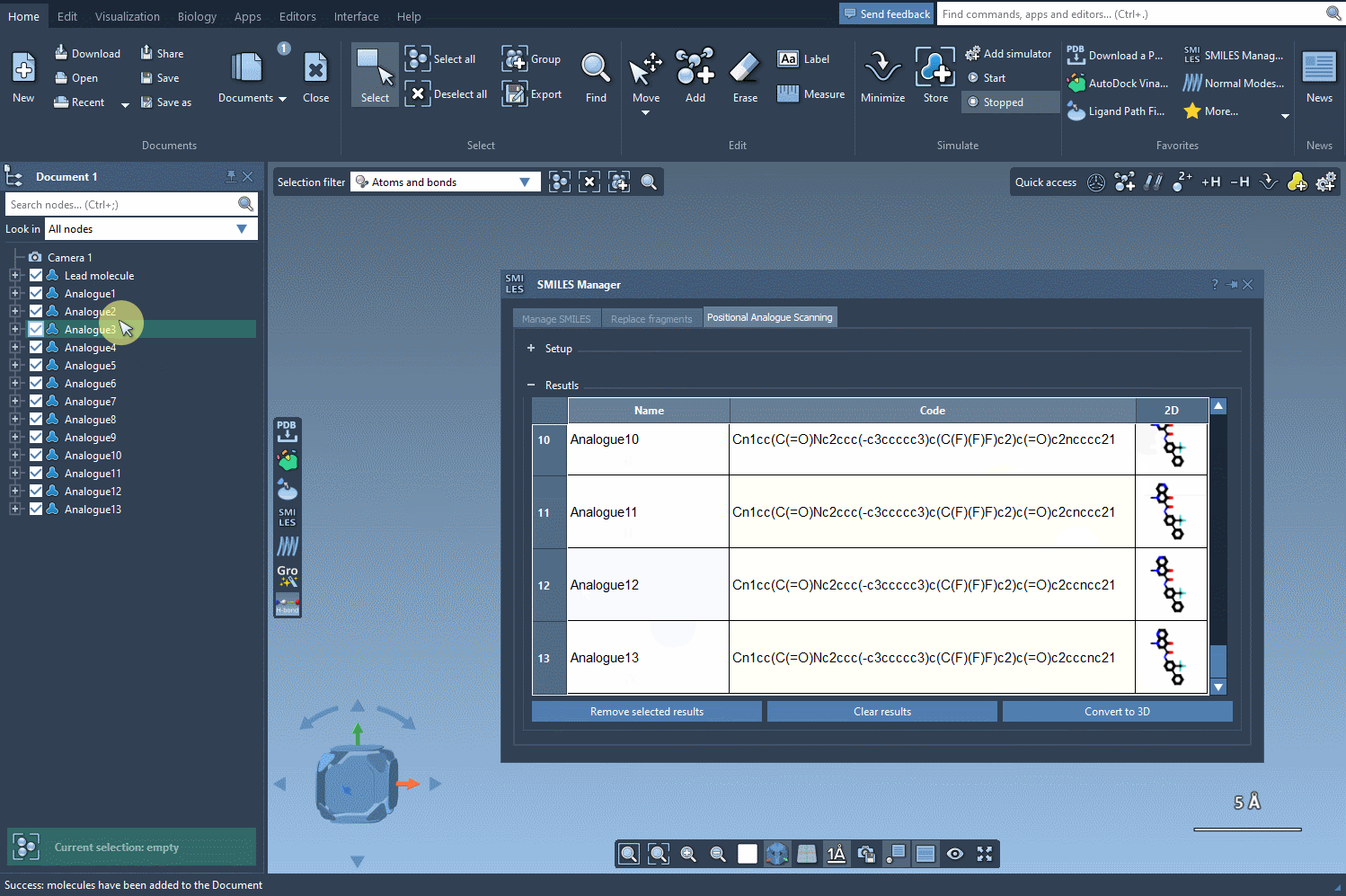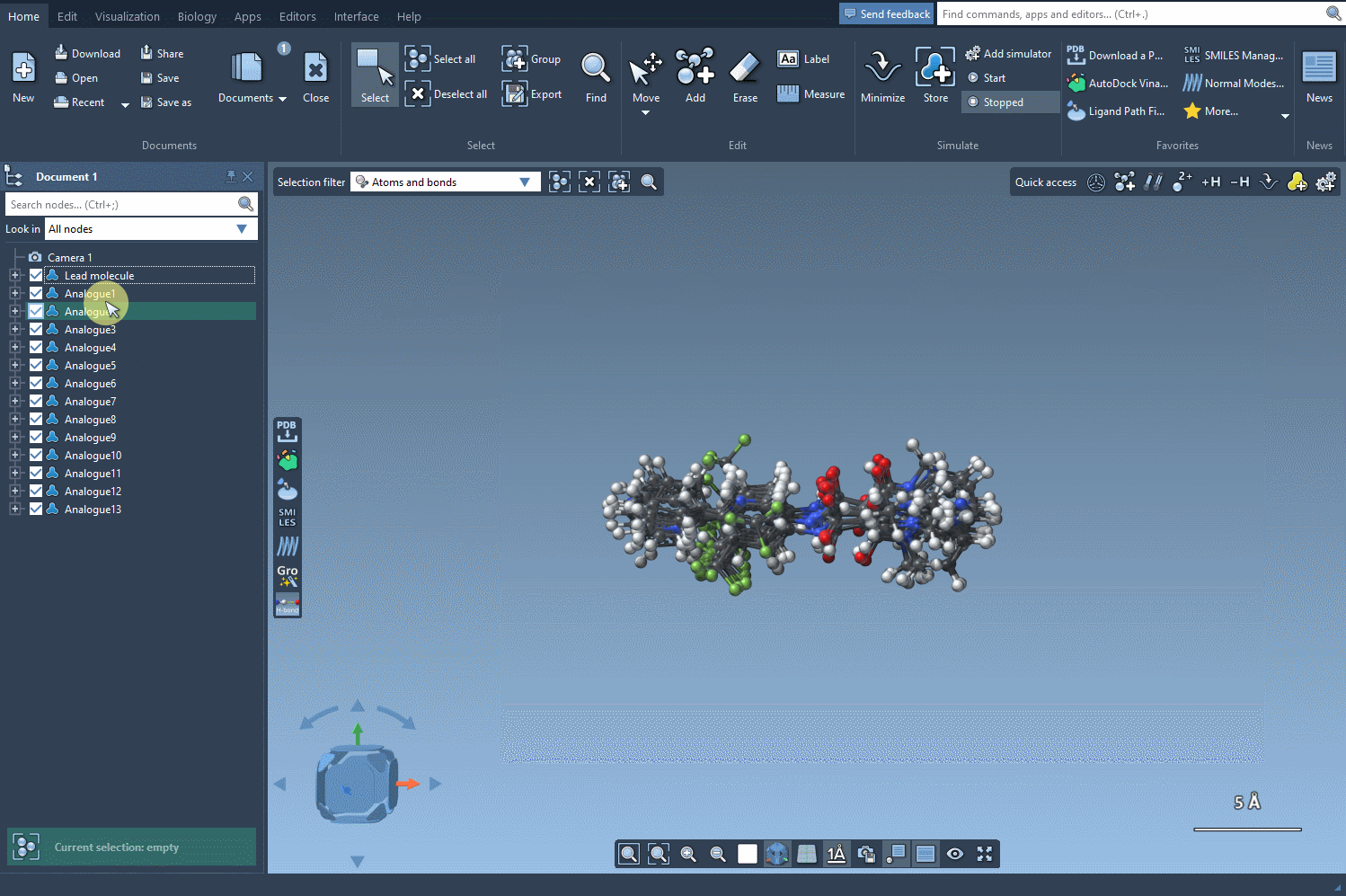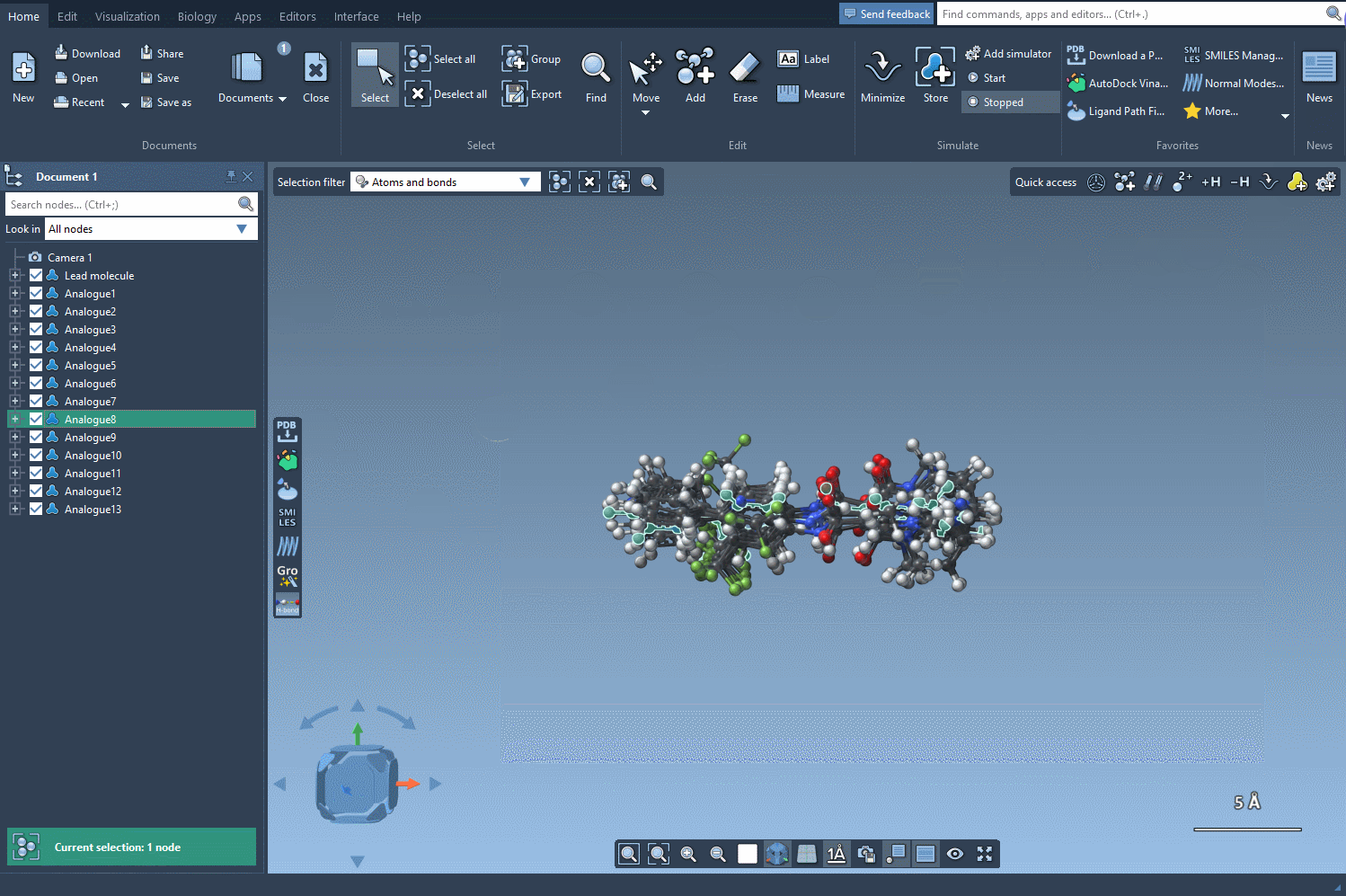
Now comes the exciting part—designing new analogs. Specify what you want to replace or attach at the highlighted positions. For example, you might decide to replace a carbon atom with a nitrogen atom (N) or attach a functional group like methyl (CH3). With a simple click of the Run button, SAMSON generates your analogs and provides immediate visual feedback, including SMILES codes and 2D depictions:
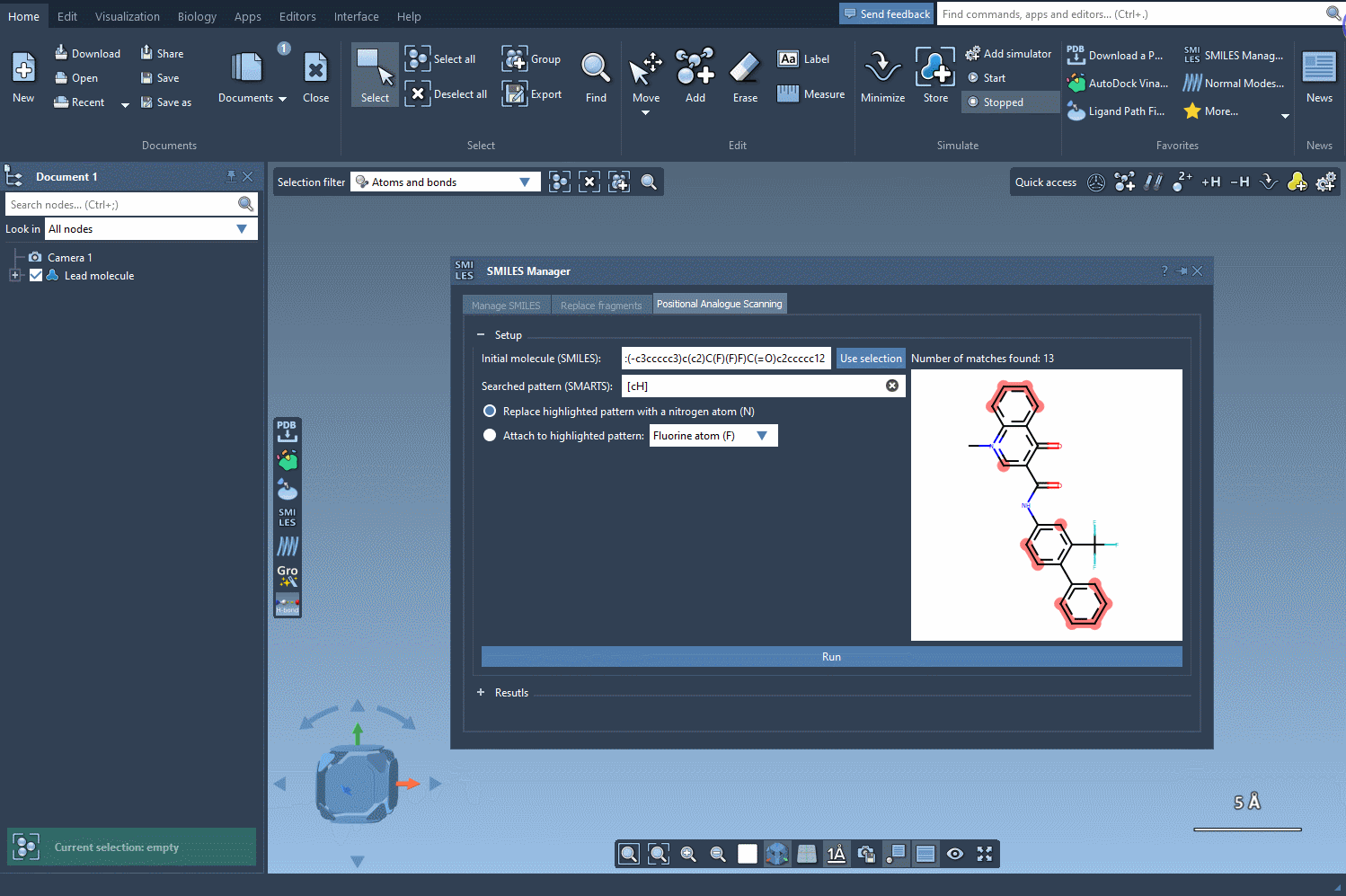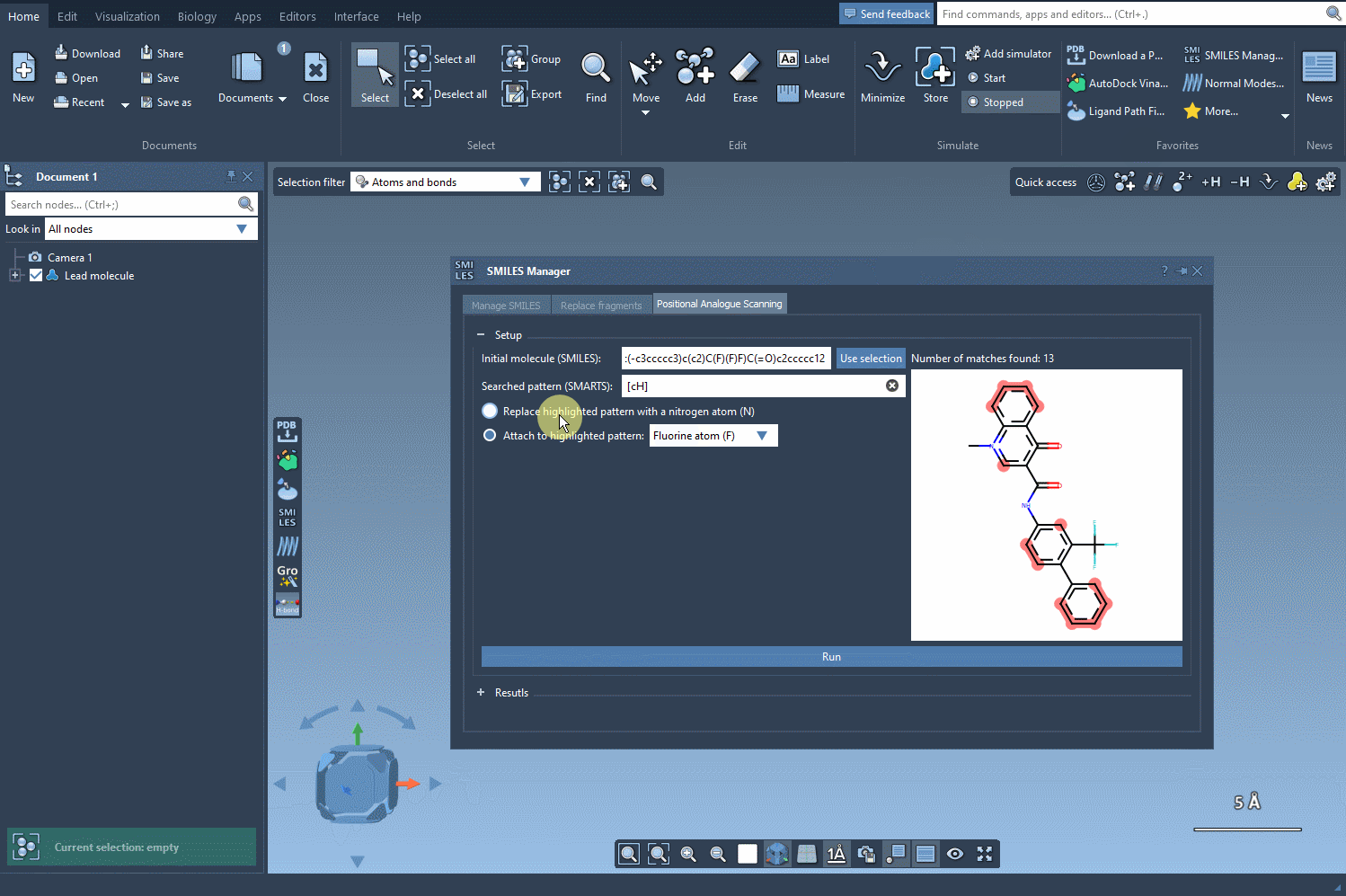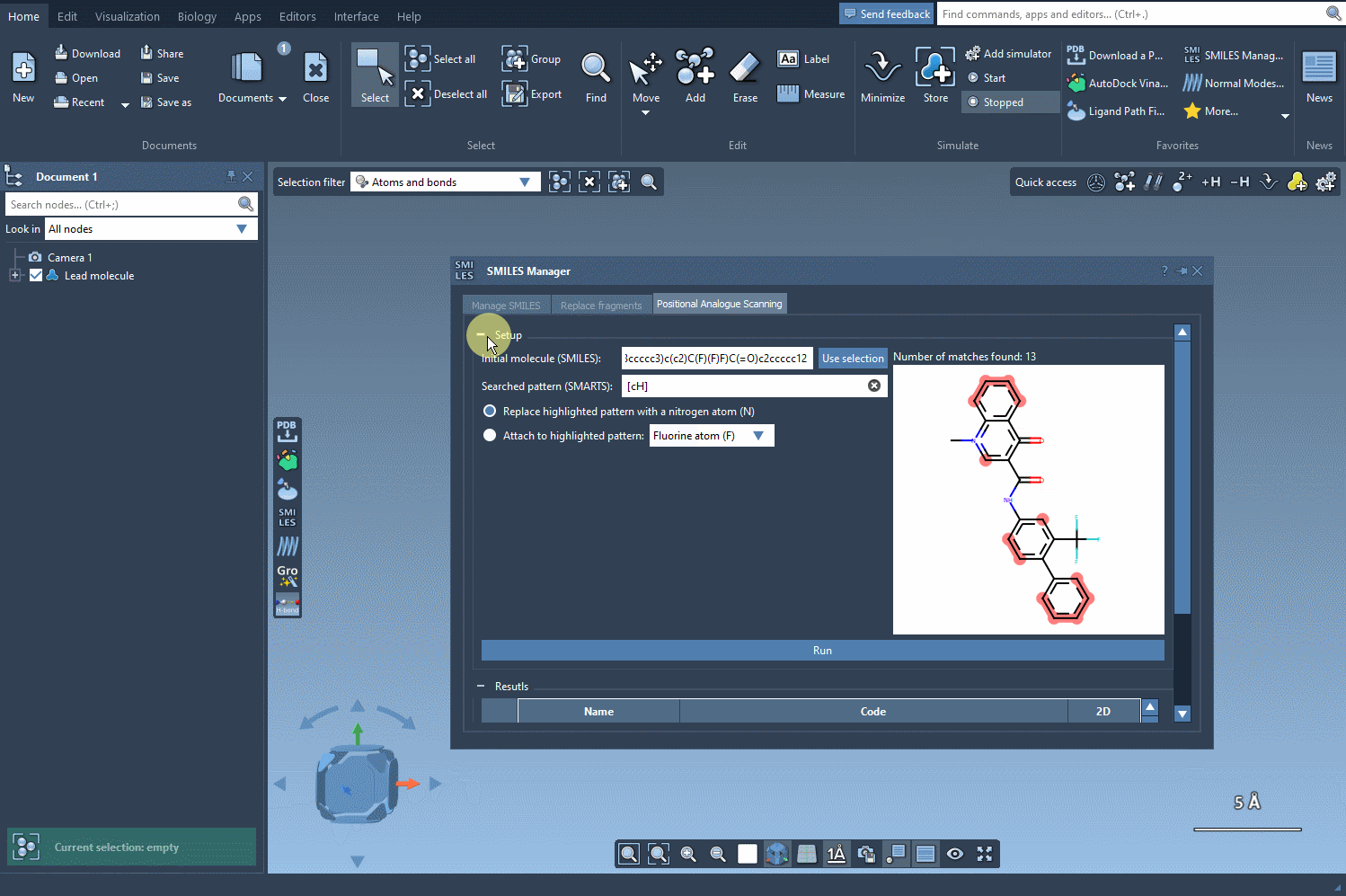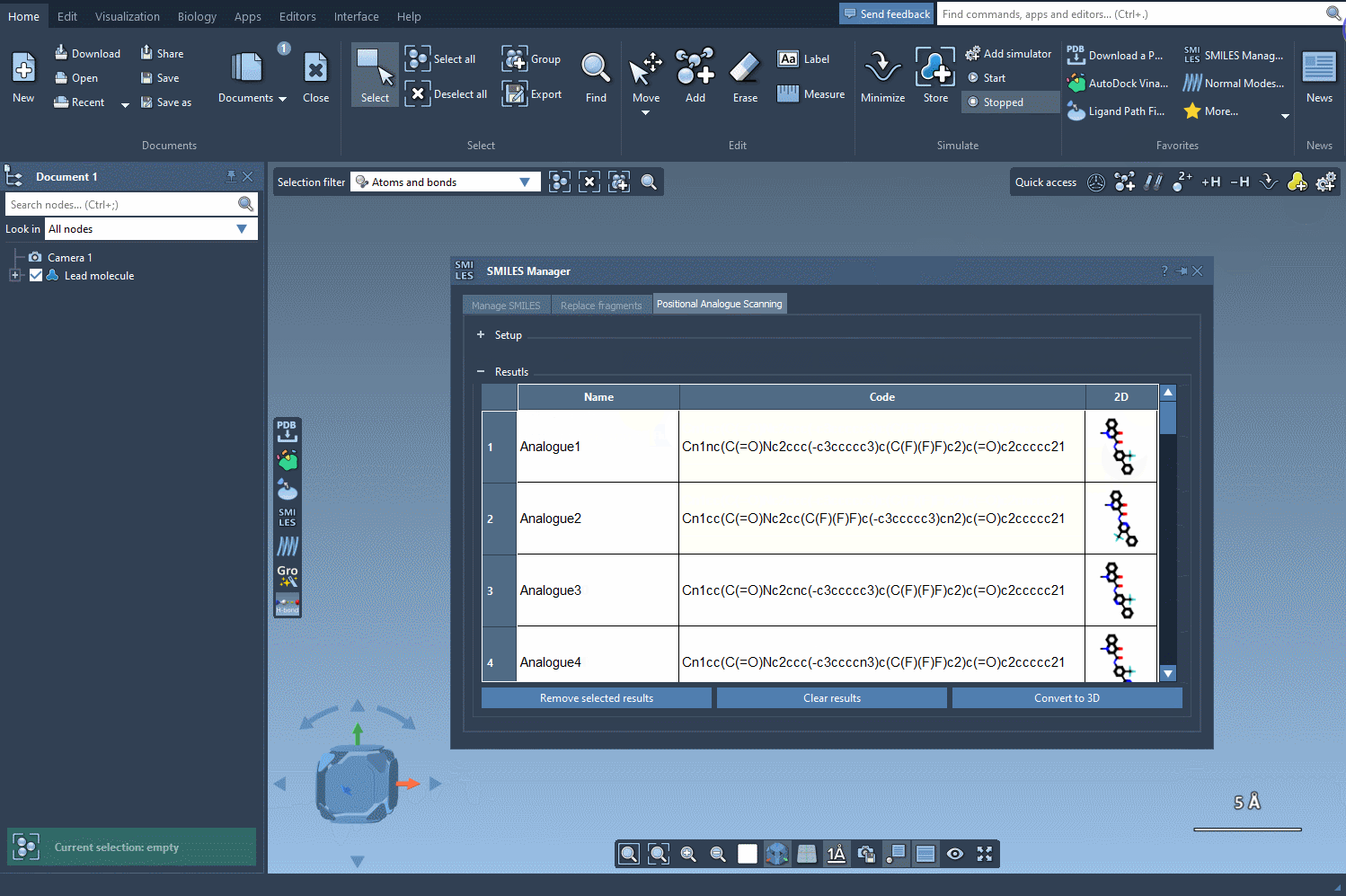
Results That You Can Refine
The results table is interactive and designed for intuitive exploration. Want to rename an analog or tweak its SMILES code? Just double-click any cell. Prefer a closer look at the generated 2D molecules? Open larger visualizations with a simple right-click. You can even generate 3D structures for specific analogs, giving you the flexibility to dive deeper into your analysis.
- Refine choices: Select multiple rows to remove unwanted analogs, or clear the entire table in one click.
- Efficient navigation: Quickly toggle the visibility of 2D images in your results to streamline comparisons.
Toward 3D Exploration
Converting your 2D results into 3D models is seamless. With just a click of the Convert to 3D button, SAMSON produces high-quality 3D structures of your analogs, ready for downstream applications like docking studies. By aligning these structures with experimental data, you can identify which analogs maintain or improve key interactions with your target protein.
Conclusion
SAMSON’s Positional Analogue Scanning tool within the SMILES Manager transforms the iterative nature of molecular design. It provides an intuitive and fast way to generate, explore, and analyze molecular analogs, offering a streamlined pathway for testing and refining new molecular ideas. To learn more about this functionality, visit the official documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. Discover SAMSON by downloading it at SAMSON Connect.





