When designing new molecules, small changes can make a big difference. Medicinal chemists often tweak functional groups or atoms to study how modifications affect binding properties, solubility, or selectivity. But making these changes manually — especially across multiple sites in a complex molecule — can be time-consuming and error-prone.
Positional Analogue Scanning (PAS) is a simple concept that becomes highly effective with the right tools. In SAMSON, the SMILES Manager extension provides a streamlined way to perform PAS: identifying parts of a molecule to modify and automatically generating analogues with atom replacements or attachments. In this post, we’ll go over how you can generate analogs with just a few clicks — and why it can help accelerate early-stage compound design.
From a molecule to dozens of analogues, fast
Let’s say you’ve got a molecule of interest and want to explore the effect of substituting certain atoms. In SAMSON, you can define this molecule by either pasting a SMILES string or selecting it directly from your SAMSON document. Once loaded, you’re ready to define the pattern to scan.
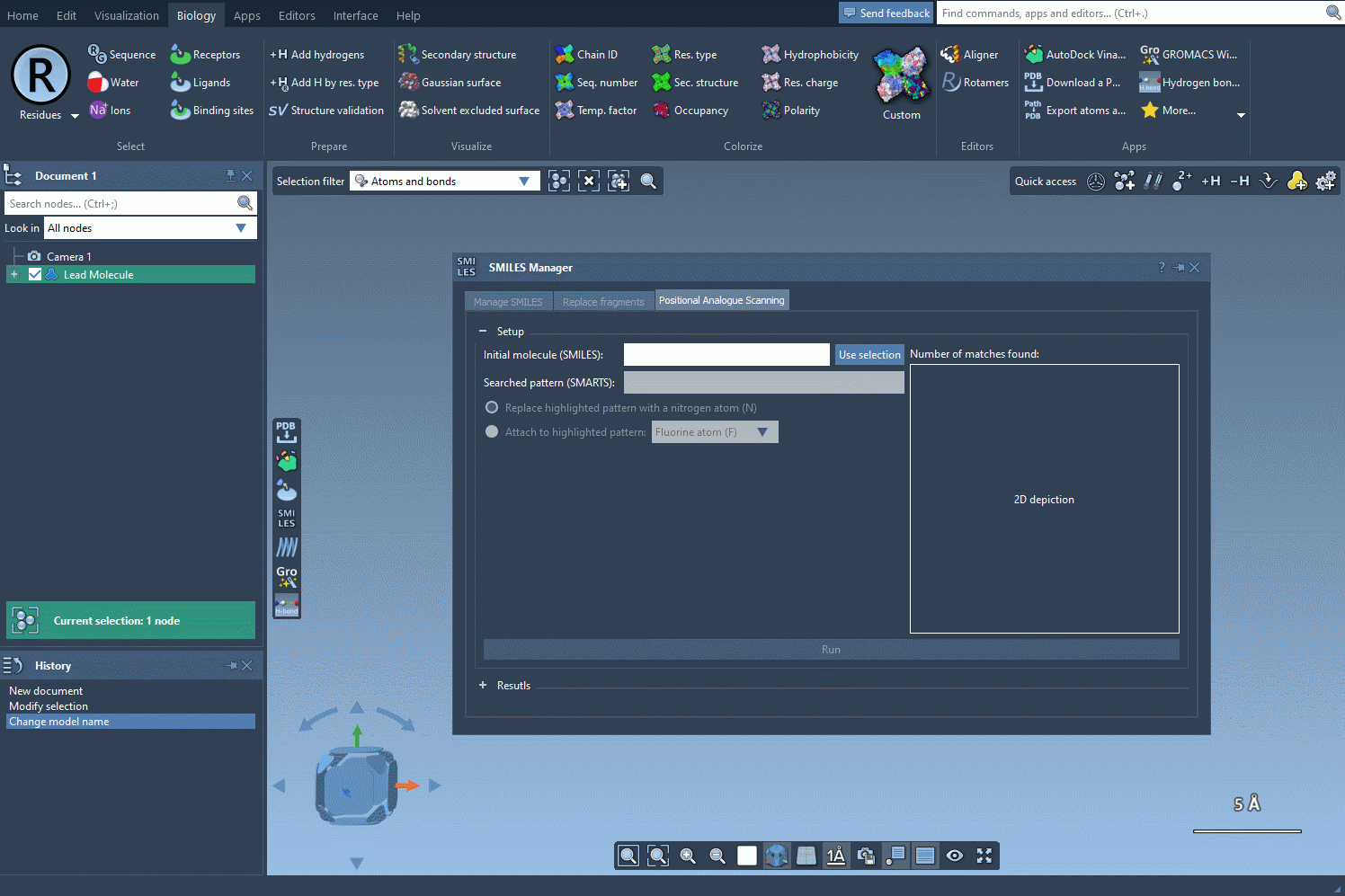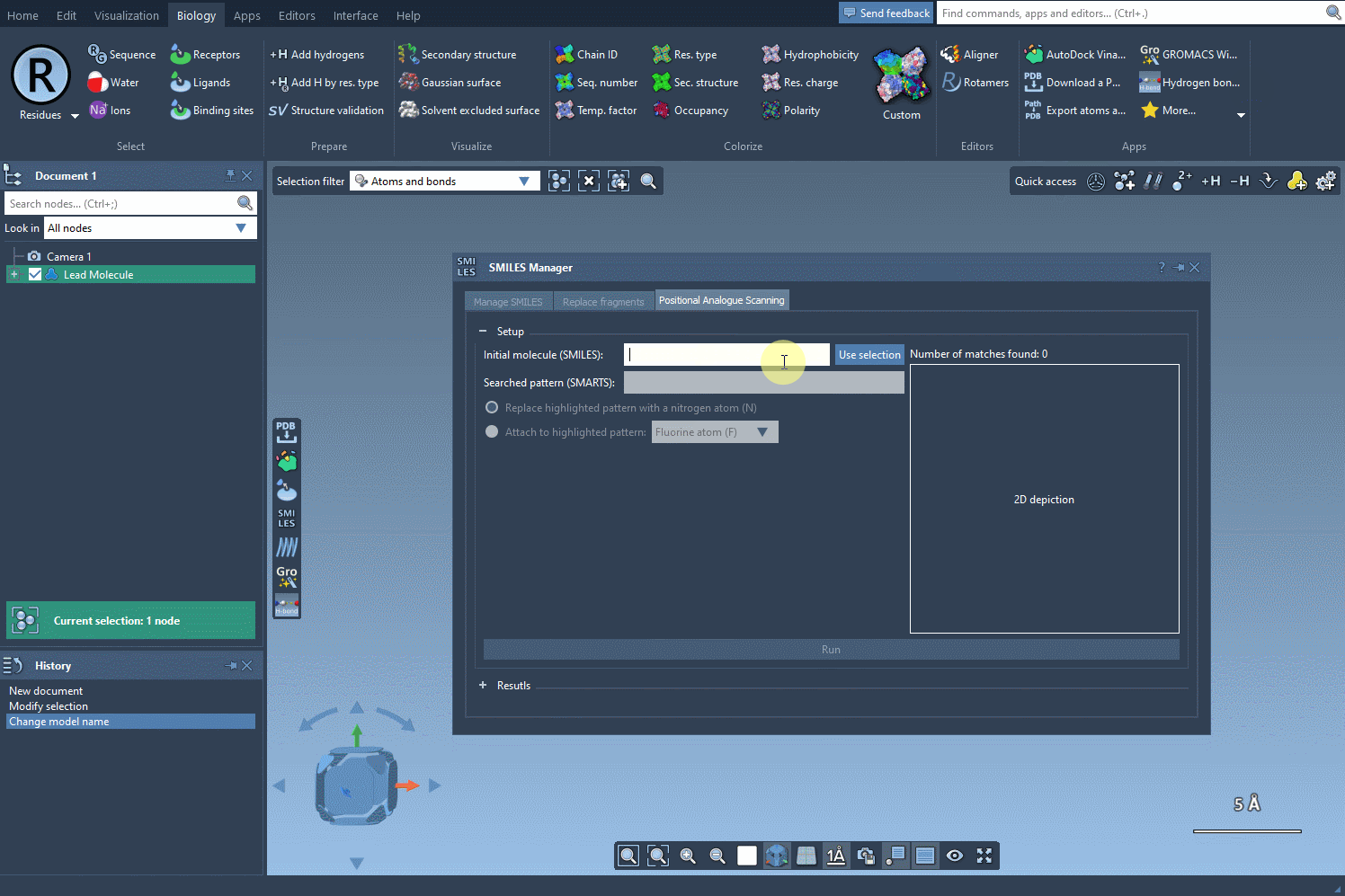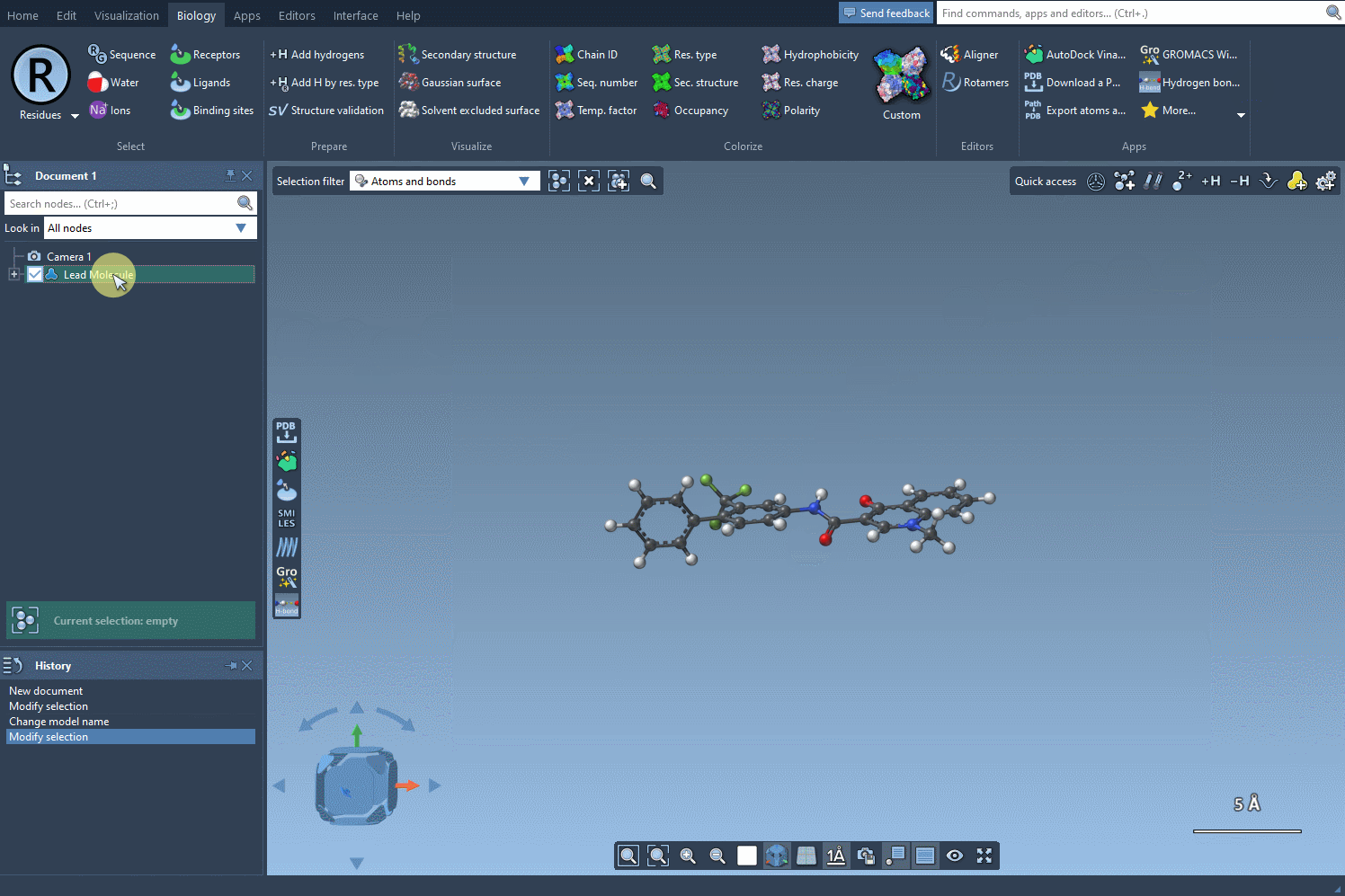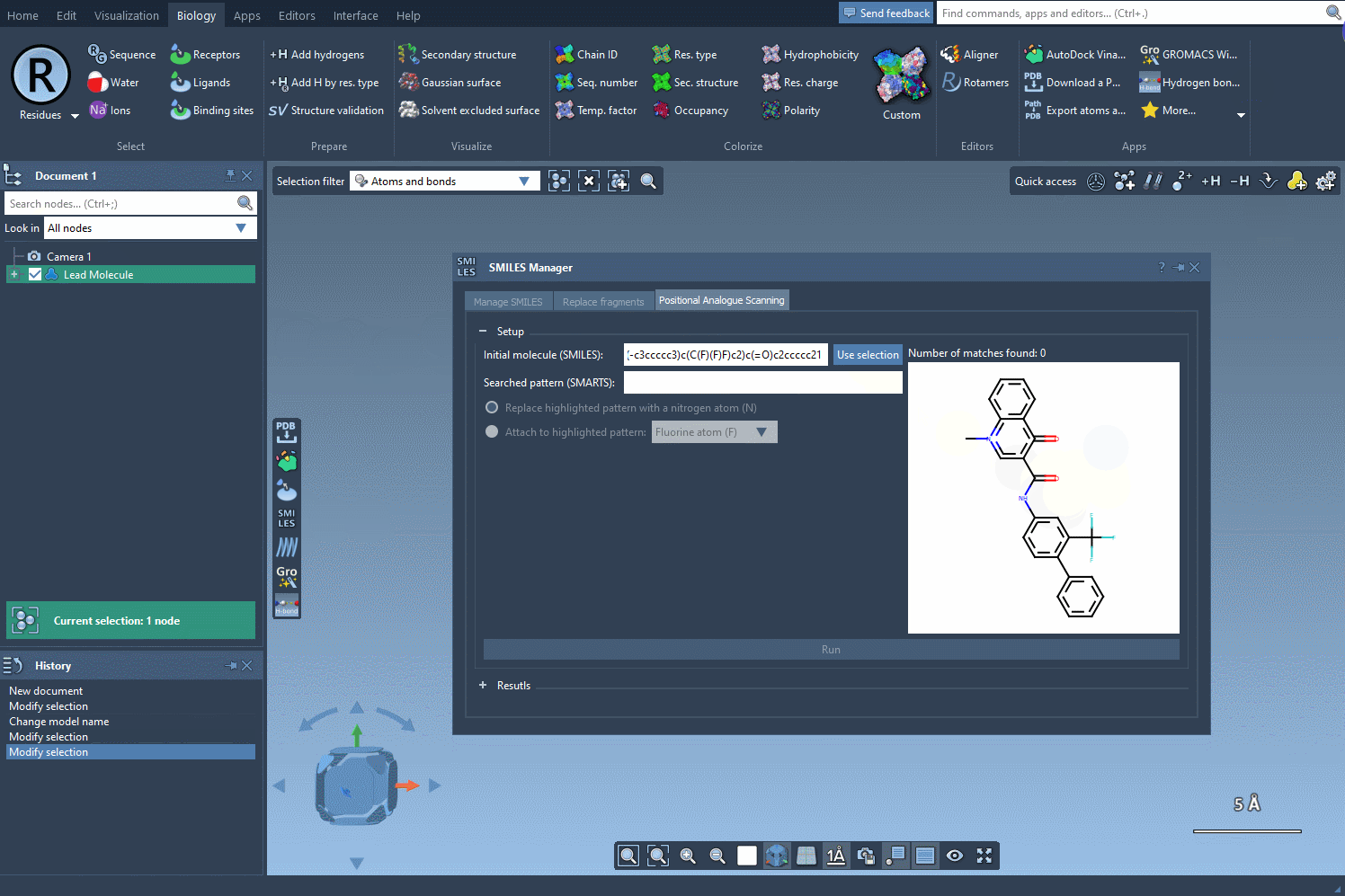
With the SMARTS language, you can target very specific atom types or configurations. For example, entering [cH] as a pattern lets you identify aromatic carbon atoms with a hydrogen — a common site for functionalization.

Replace or attach new groups
Once you’ve defined your search pattern, the real power of the SMILES Manager kicks in. You can:
- Replace the matched atom with another (e.g., carbon with nitrogen)
- Attach a group to the matched atom (e.g., adding a fluorine or methyl group)
Click Run, and the extension will generate all possible derivatives at each matching site. It’s a fast way to explore single-point mutations on a molecule without drawing them one by one.
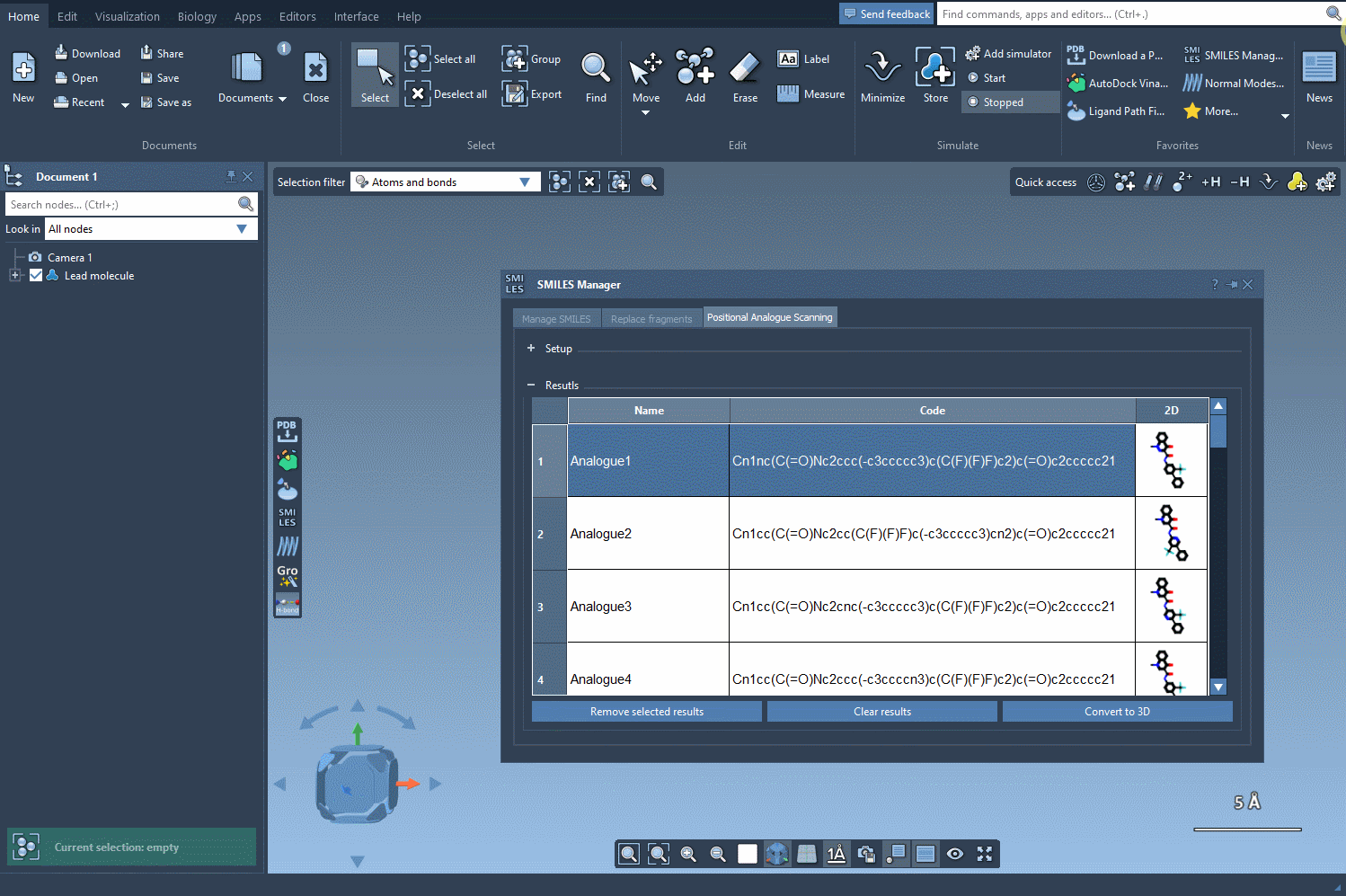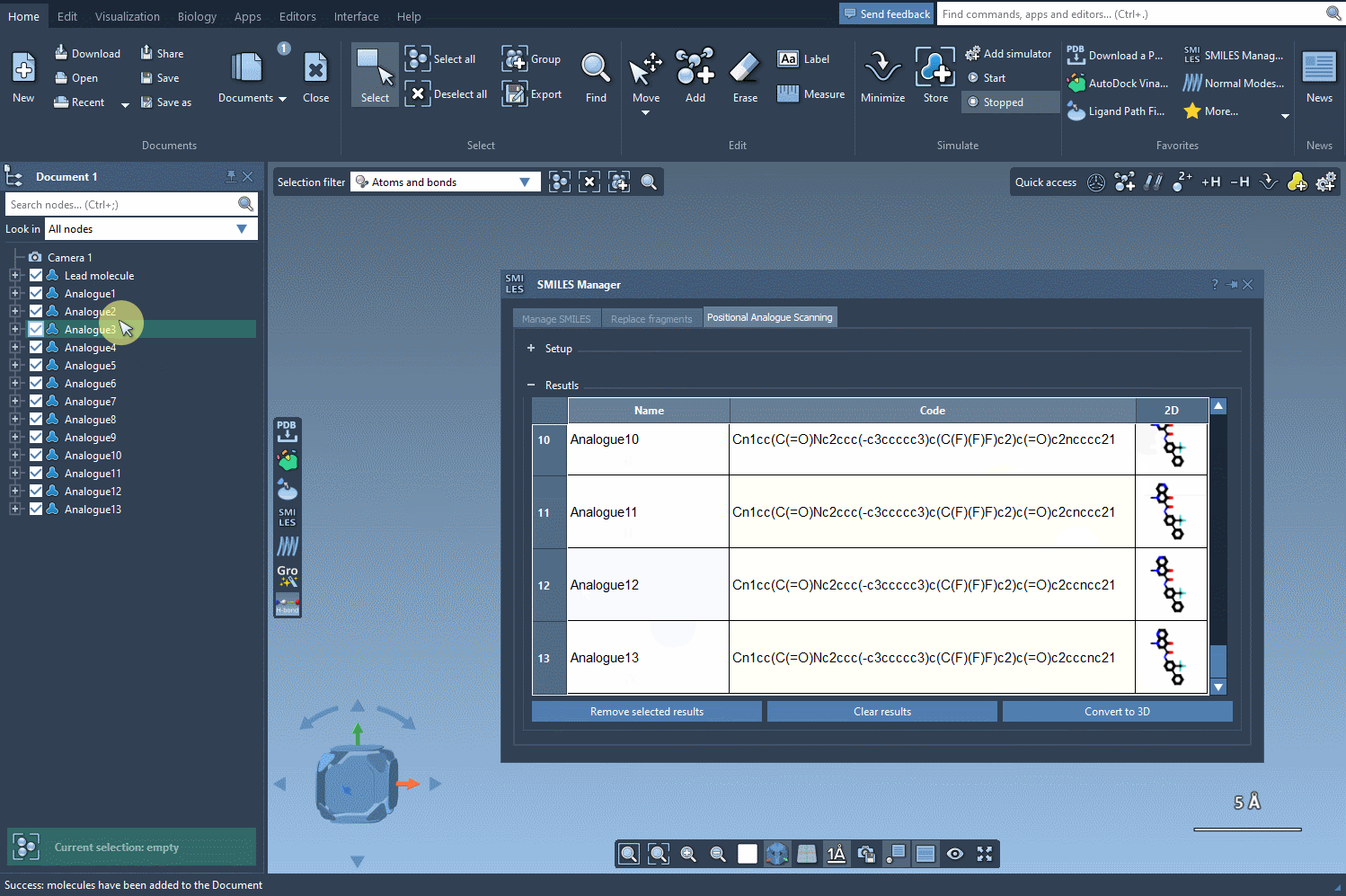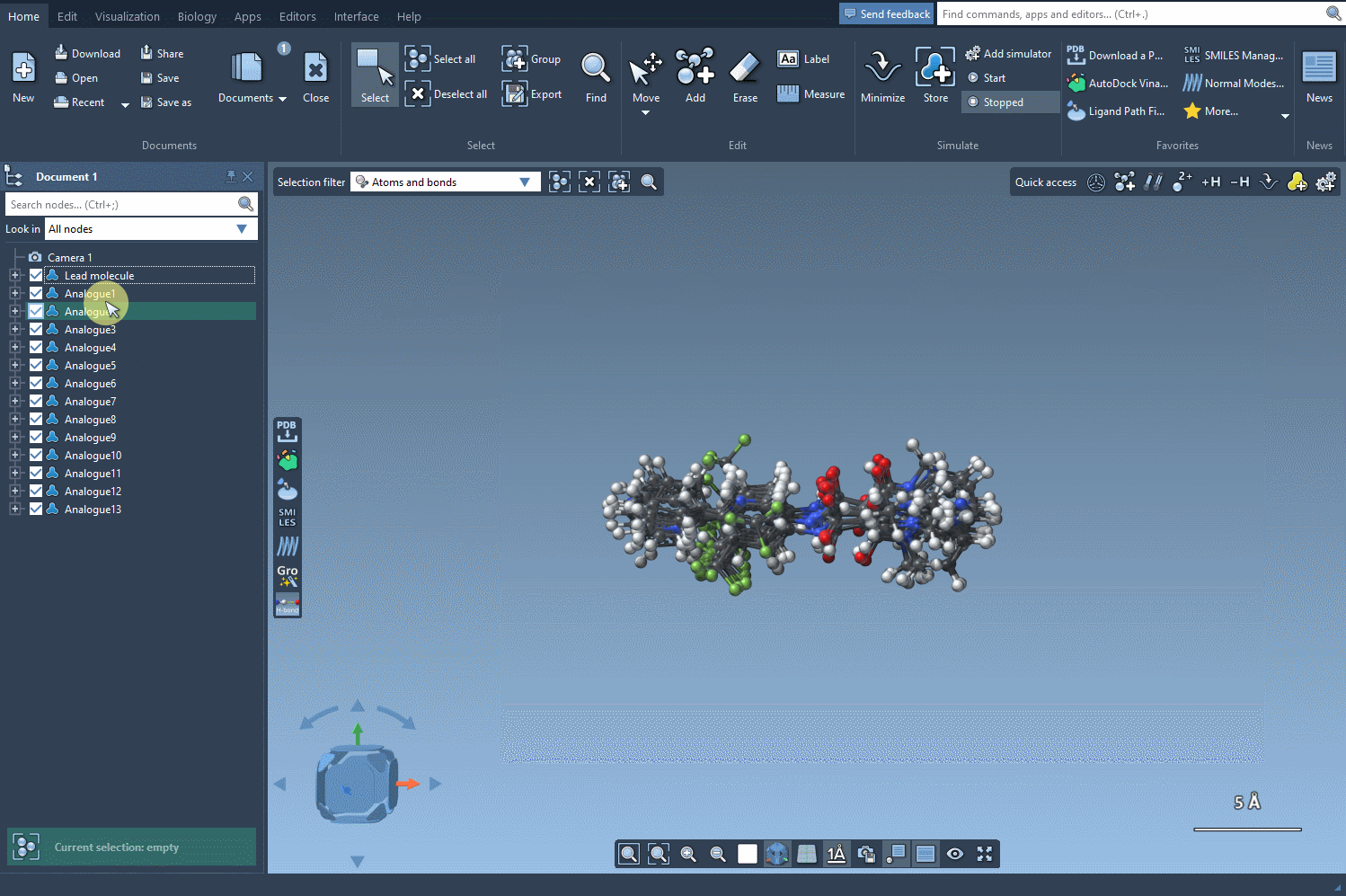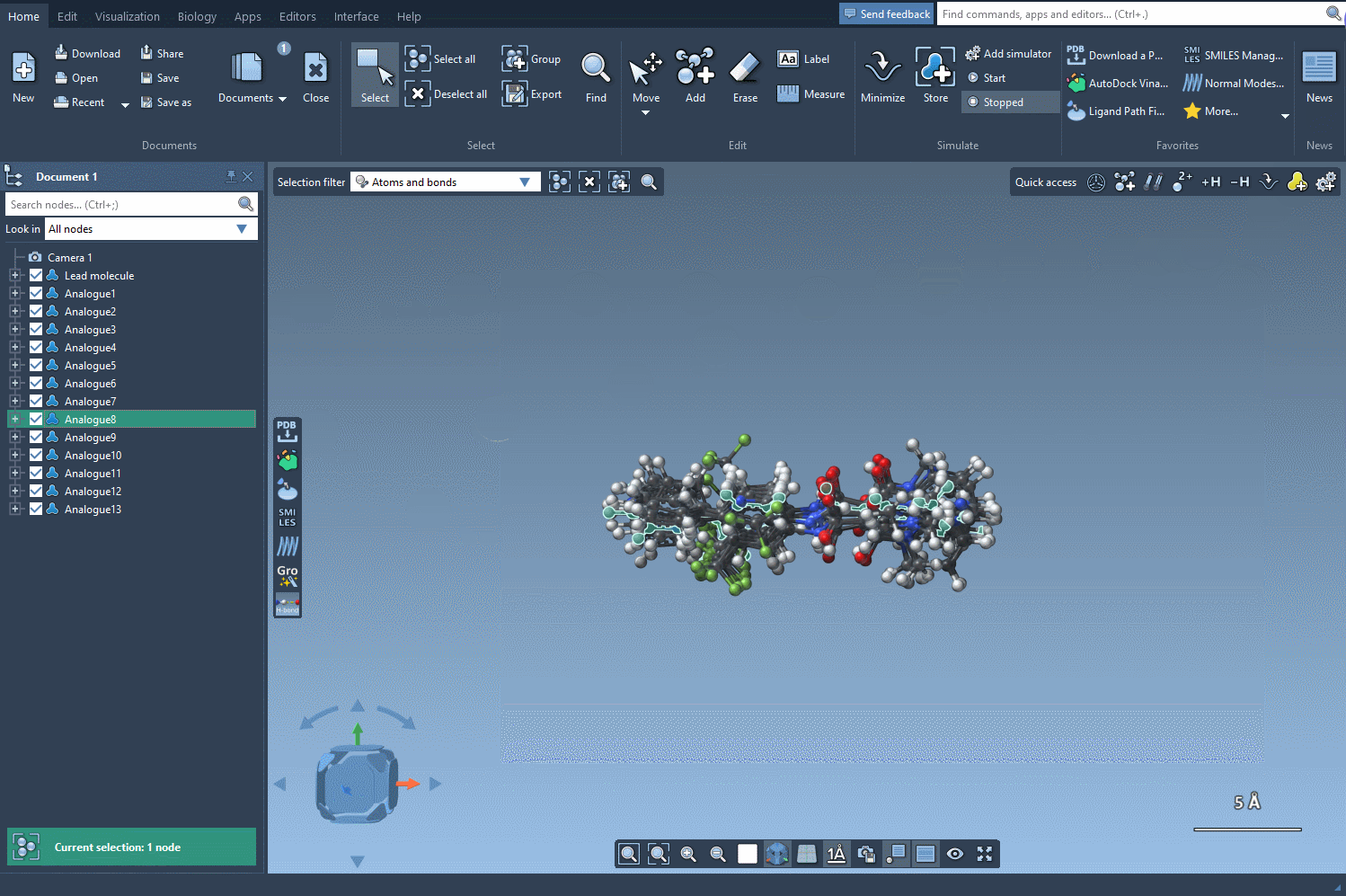
View and adjust results
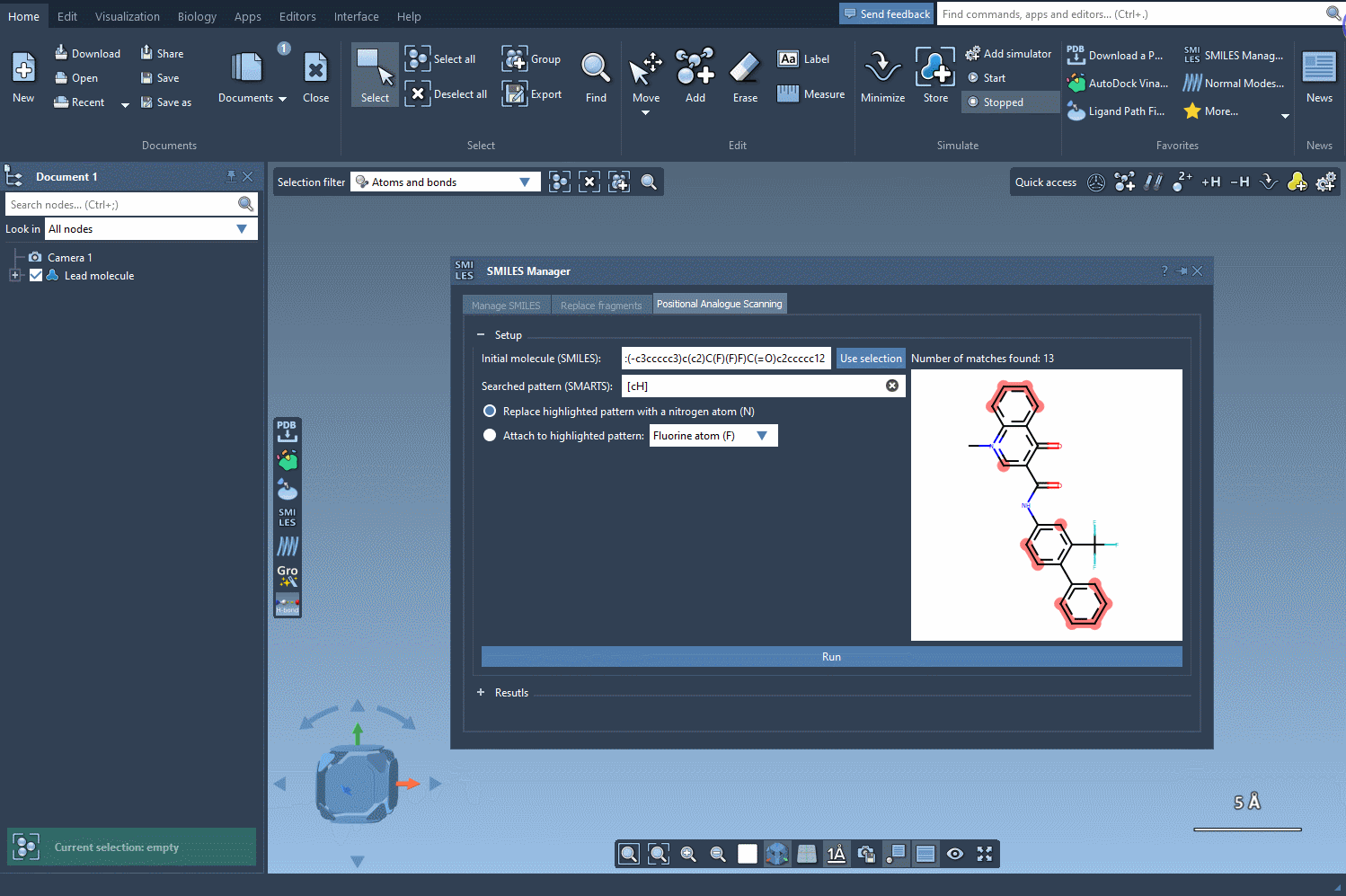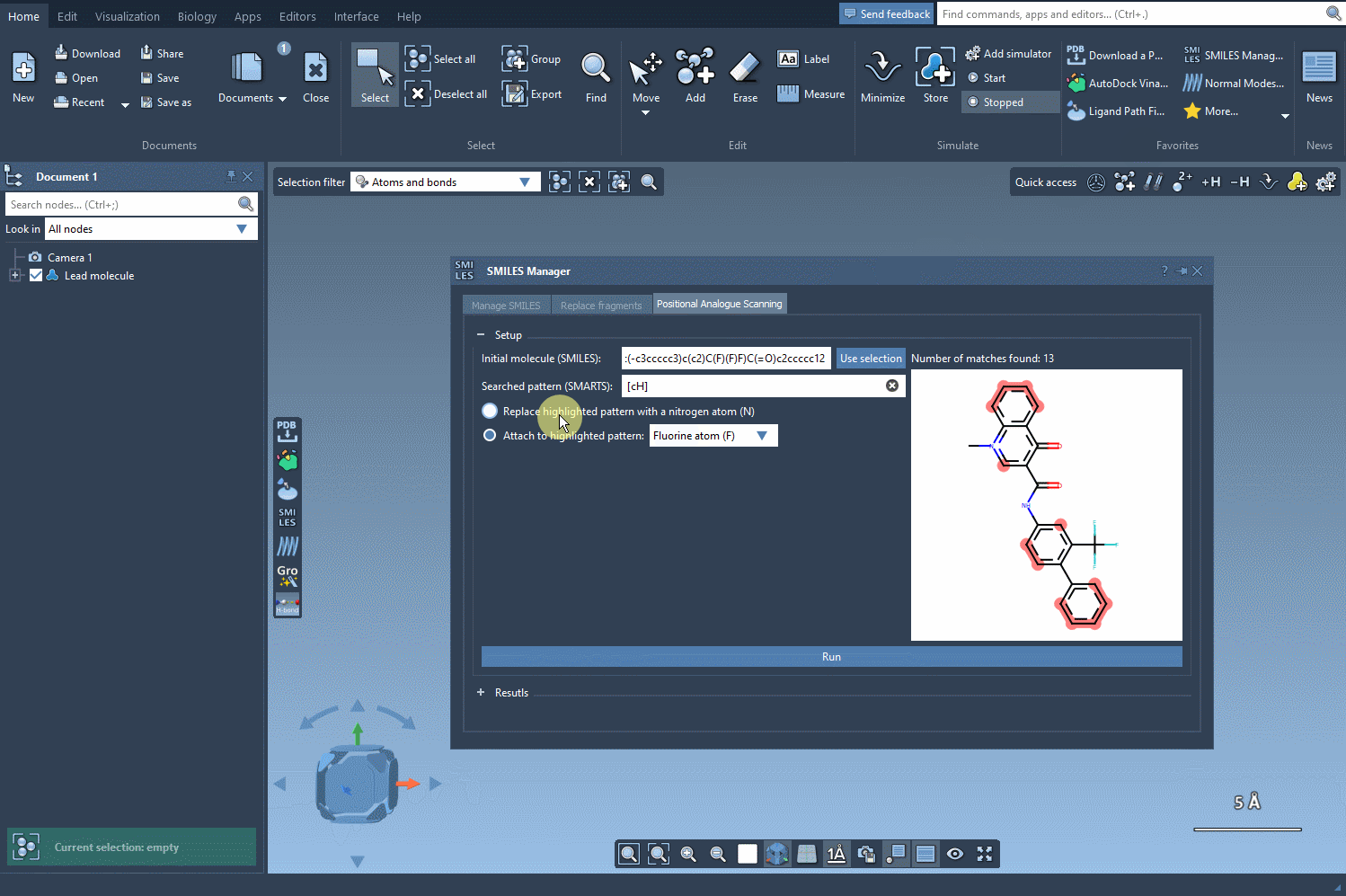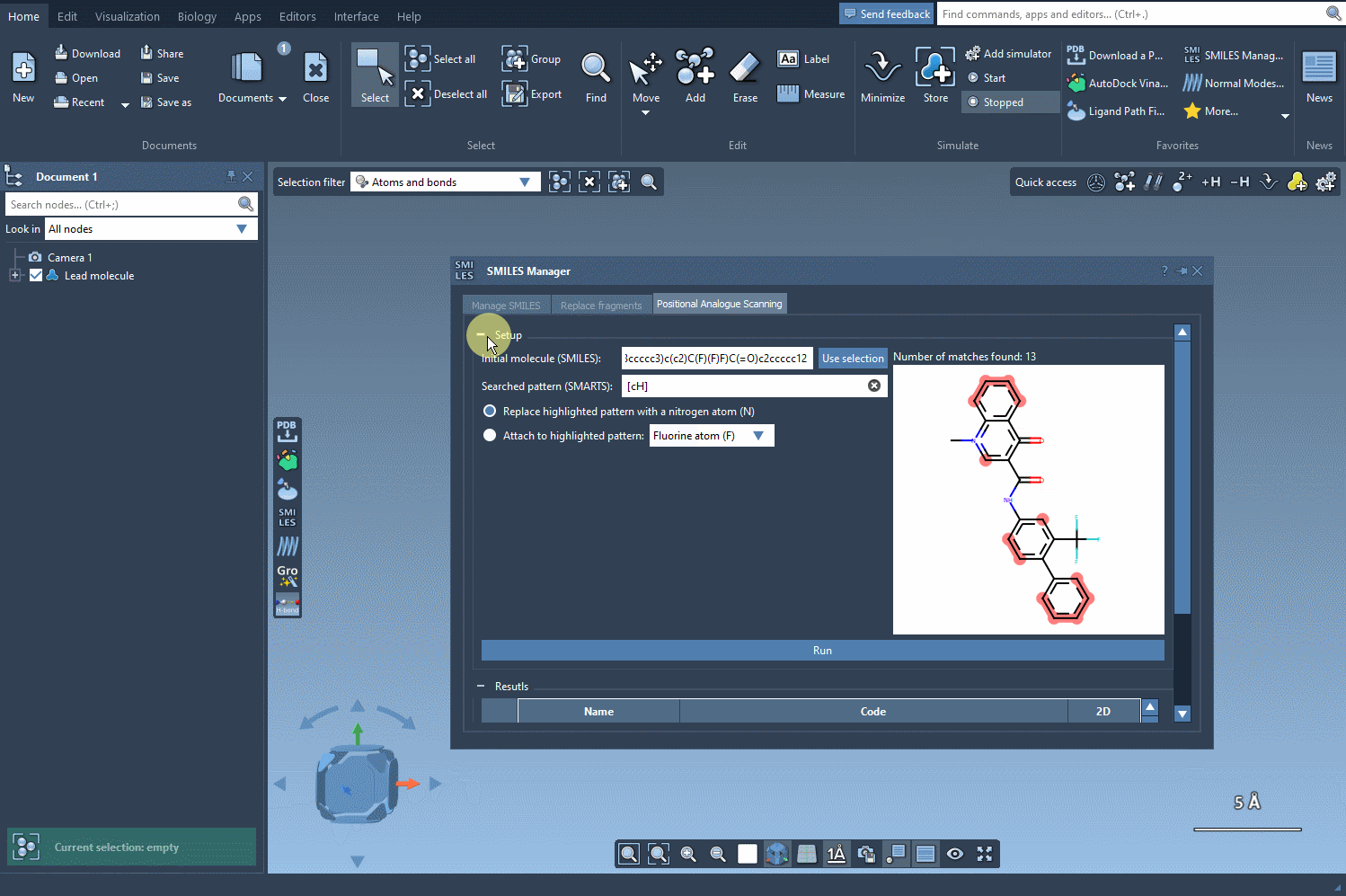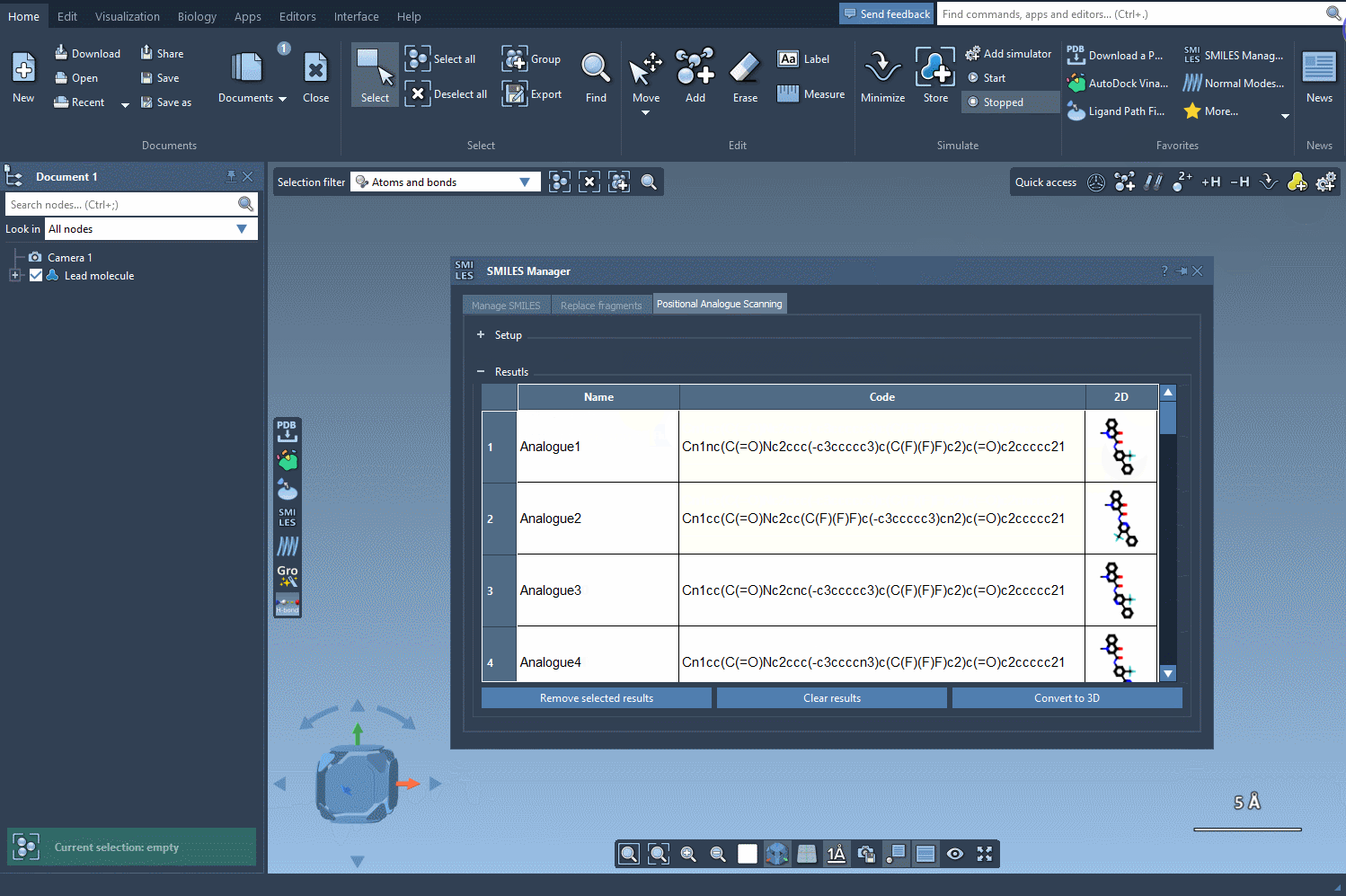
The results appear in an interactive table displaying SMILES, 2D depictions, and more. You can edit names or structures, inspect 2D images, or remove certain analogs. Then, for deeper analysis, select any analogue and generate a 3D model directly from its SMILES with one click.
Why it matters
Systematically generating analogues is a key part of structure-activity relationship (SAR) studies. Small modifications help uncover which molecular features are responsible for activity — or lack thereof. With PAS and the SMILES Manager in SAMSON, this step becomes tractable even for large datasets. You’re not repeating steps manually. You’re scanning possibilities with precision and speed.
You can even follow up immediately by assessing binding via docking simulations (with tools like Autodock Vina Extended), or visual interaction analysis, all within the same environment.
If you’re working in early-stage drug design or just curious to explore chemical space more efficiently, this is a workflow worth knowing.
To learn more, check the full tutorial documentation on Performing Positional Analogue Scanning using the SMILES Manager.
SAMSON and all SAMSON Extensions are free for non-commercial use. Get SAMSON at https://www.samson-connect.net.





