Setting up molecular simulations can be frustratingly complex, especially when you’re trying to model systems that undergo structural changes. In many tools, altering bonds or atom types mid-simulation requires stopping, resetting, and restarting the entire process. That’s a workflow killer. 😓
This is where IM-UFF—the Interactive Modeling Universal Force Field—comes in. It’s a powerful extension to the well-known UFF (Universal Force Field) that lets you model molecular systems where the topology isn’t fixed. Let’s walk through how to quickly configure IM-UFF in SAMSON, and see how simulation work can happen more naturally—with fewer interruptions, and greater flexibility.
When Topology Won’t Stay Still
One of the most common pain points in molecular modeling is dealing with systems under transformation: reactions where bonds break or form, or where atom types need to be assigned on the fly. Standard force fields need a predefined topology and don’t adapt well to these changes. The IM-UFF model addresses this precisely—it updates molecular structures as you work with them interactively, allowing topologies to evolve throughout a simulation.
Setting Up IM-UFF
You only need a few steps to enable interactive molecular modeling:
- Open your molecular system in SAMSON.
- Navigate to
Edit > Simulate > Add simulator(or useCtrl+Shift+M/Cmd+Shift+M). - Select Interactive Modeling Universal Force Field from the list of interaction models.
- Choose a simulation method like FIRE (Fast Inertial Relaxation Engine).
- Click OK.
Now the magic begins! You’ll see IM-UFF’s parameter window. Unlike standard UFF, there’s no separate setup window to go through, keeping the workflow smooth.
Two Options Worth Knowing
IM-UFF offers two configuration options in its parameter window that directly relate to modeling flexibility:
- Static topology (UFF only): Check this if you want to freeze the topology (like traditional UFF). Uncheck it to enable topology changes using IM-UFF.
- Keep vdW for manipulated: When you’re dragging atoms, the van der Waals forces can hinder motions due to repulsion. Unchecking this option ignores vdW for moved atoms, making it easier to guide them into new positions.
Once your simulator is set and the options configured, click Edit > Simulate > Start. You’re now simulating with IM-UFF.
What Happens During Simulation?
Interactively manipulating atoms gives a more intuitive way to model systems. Small moves will preserve bonds and the system adjusts nearby atoms accordingly. Larger displacements can break bonds and trigger atom retyping and bond reassignments dynamically, reflecting real chemistry behaviors.
Here’s a live view of an interactive simulation in action 👇
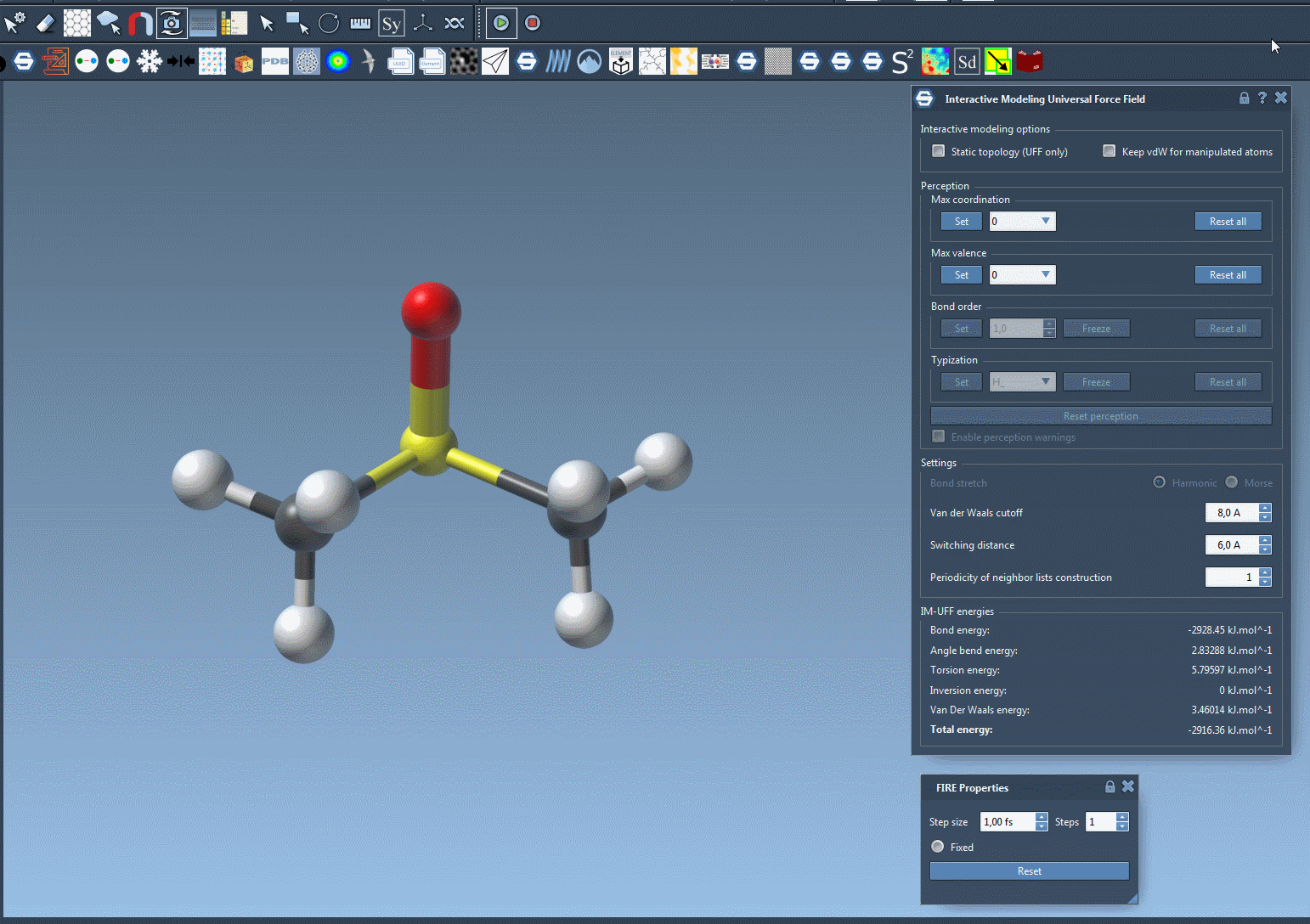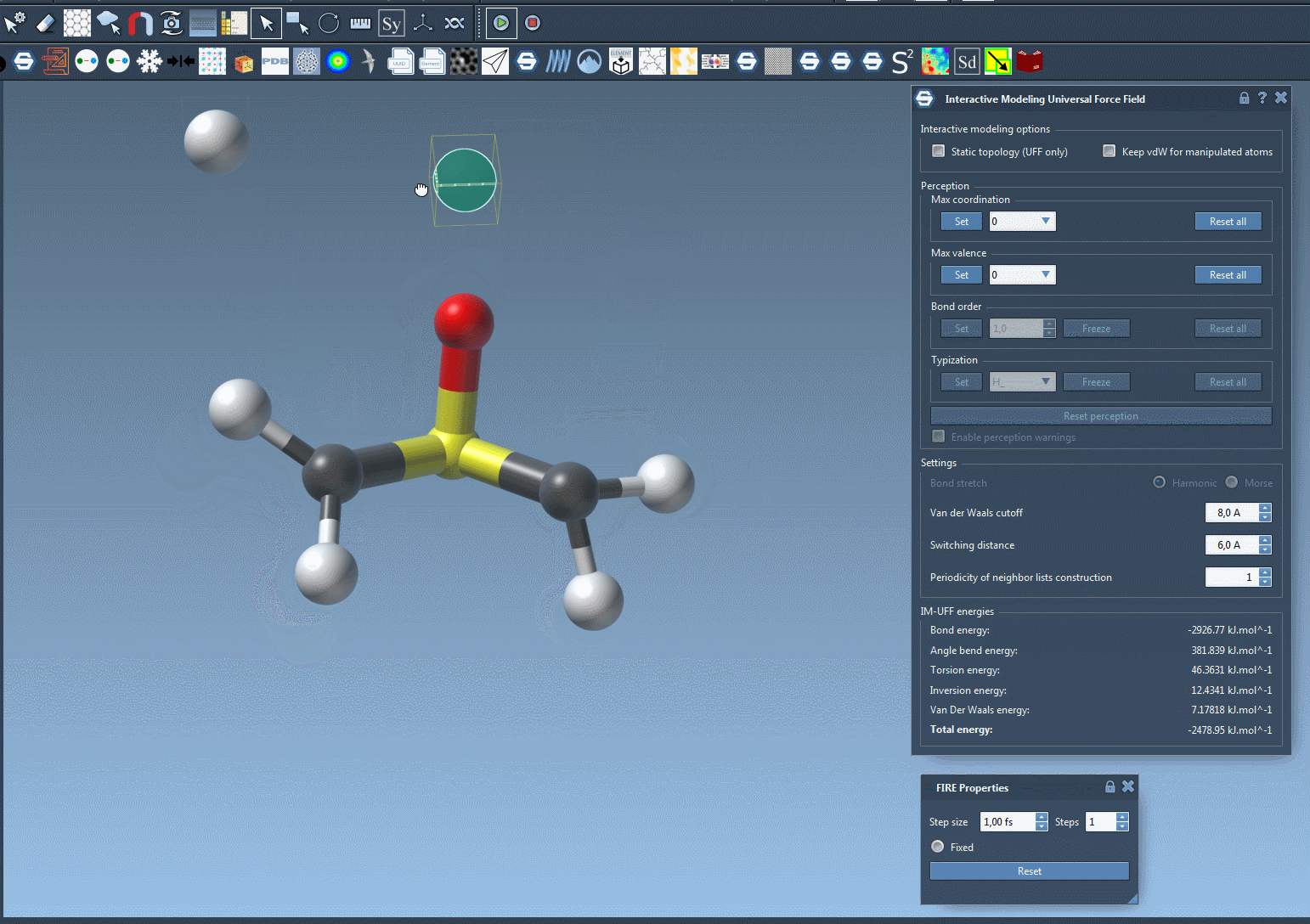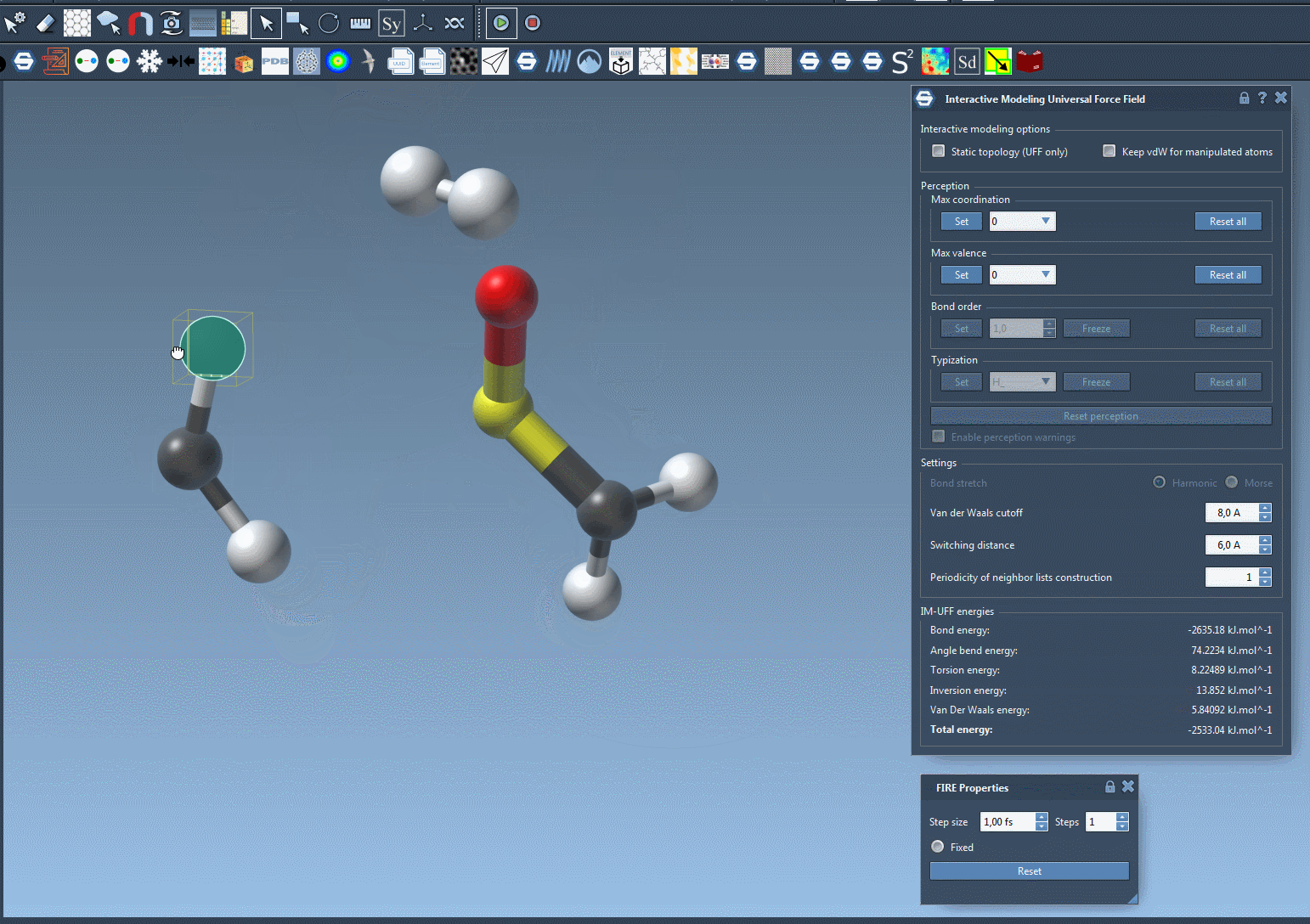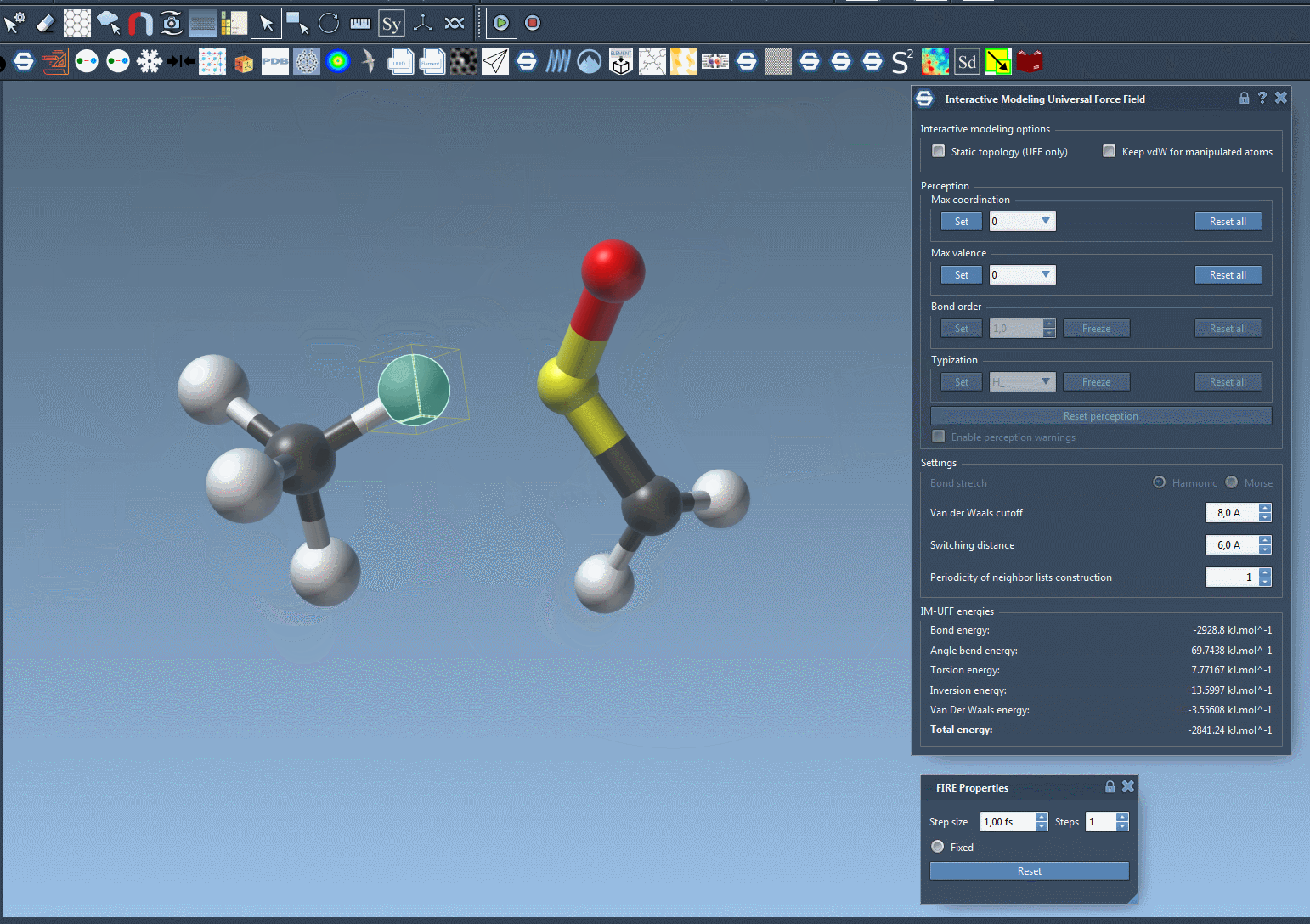
This kind of fluid modeling process allows you to build or repair topologies as you simulate, making it ideal for molecular design workflows that don’t follow a rigid, predefined pathway.
To Learn More
Setting up simulations with evolving topologies doesn’t have to be hard. IM-UFF provides an adaptive approach that greatly reduces interruptions and improves realism in molecular modeling.
Read the full documentation for more detailed instructions and customization tips.
SAMSON and all SAMSON Extensions are free for non-commercial use. Get SAMSON at https://www.samson-connect.net





