One of the more challenging tasks in molecular modeling is understanding how a ligand navigates its way out of a protein binding pocket. Whether you’re studying drug binding kinetics, escape routes, or trying to visualize molecular flexibility, it can be hard to track movements in complex systems.
If you’ve ever found yourself asking, “How do I follow where the ligand goes during a simulation, especially among all those atoms?”—you’re not alone.
The Pathlines extension in SAMSON provides a visual and intuitive way to track center-of-mass (COM) movements of selected atom groups—ideal for observing ligand unbinding and conformational changes. Here’s a walk-through of how you can visualize ligand escape paths with COM pathlines using the provided Lactose permease sample system.
Visualizing Ligand COM Trajectories
The tutorial sample system contains 1PV7 bound to Thiodigalactosid (TDG) along with unbinding paths generated using the Ligand Path Finder app. Instead of manually tracing every atomic detail, you can visualize the movement of the ligand’s center of mass along these precomputed paths.

Step-by-Step Guide:
- Select the ligand: In the Document view, click on the TDG molecule. (Hold Ctrl/Cmd to select multiple components if needed.)
- Select the paths: Choose one or more of the unbinding paths shown in the sample.
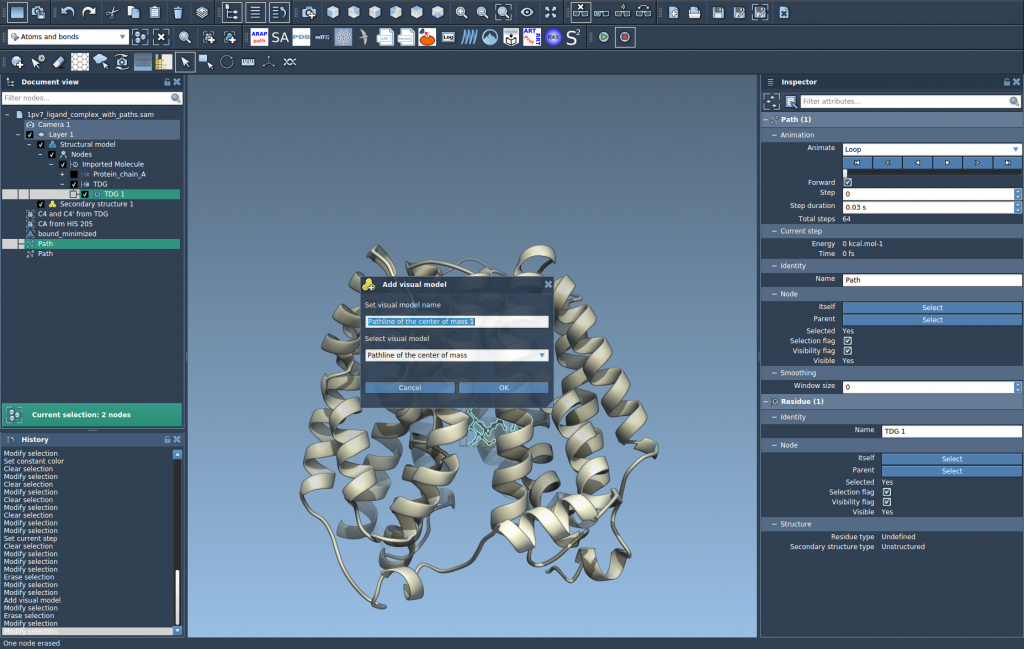
- Create a visual model: Go to
Visualization > Visual model > More...and select Pathline of the center of mass.
This will generate a visual curve representing how the COM of the ligand moves along the specified path(s).
Inspect and Fine-Tune the Visualization
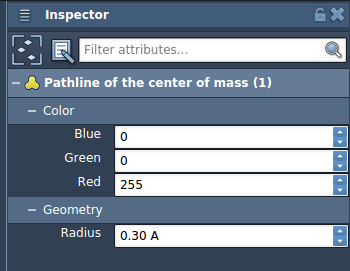
Once the pathline is created, you can customize its appearance by selecting it and opening the Inspector (Ctrl/Cmd + 2). You can adjust color, line thickness, and other visual properties for better clarity.
This is particularly helpful when visualizing multiple ligands or comparing different escape paths. You can even animate the path over time or overlay multiple pathlines.
Why It Matters
Classic root-mean-square deviation (RMSD) plots or coordinate logs only tell part of the story. A clear, spatial representation of movement helps understand kinetics, diffusion, and mechanistic pathways. Using COM-based pathlines streamlines this process, eliminating visual noise while keeping high fidelity for motion analysis.
This is especially practical when working with unbinding simulations, steered molecular dynamics, or reaction coordinate studies.
Ready to Try It?
You can start using pathline visualizations by first visiting the Pathlines Extension Page and adding it. Then follow the steps above using the provided Lactose permease sample system.
To learn more, read the full documentation at: https://documentation.samson-connect.net/tutorials/pathlines/pathlines/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





