One recurring challenge in structural biology and molecular modeling is how to study conformational changes between two known protein structures. Whether you’re investigating molecular mechanisms, setting up umbrella sampling simulations, or simply trying to better understand flexibility in biomolecules, capturing a plausible intermediate transition path can be both essential and time-consuming.
Fortunately, SAMSON’s ARAP Interpolator extension introduces a fast and intuitive way to animate transitions between two protein conformations using As-Rigid-As-Possible (ARAP) interpolation. Here’s how you can generate a smooth structural animation — and why you might want to do it in the first place.
Why Animate Protein Transitions?
Proteins are not static. Their function often depends on large-scale motions — for example, the opening and closing of an enzyme’s active site or the rearrangement of domains upon ligand binding. Visualizing these motion trajectories allows researchers to:
- Generate hypotheses about mechanistic pathways
- Set up path-based simulations like umbrella sampling or steered MD
- Improve understanding when communicating structural insights
Using ARAP for Protein Transition Paths
Inspired by deformation modeling in computer graphics, ARAP interpolation attempts to preserve local geometry (i.e., be “as rigid as possible”) while interpolating atom positions. This keeps the motion realistic without requiring a full molecular dynamics simulation.
To get started, install the ARAP Interpolator Extension from SAMSON Connect. Once installed, you can launch the app from Home > Apps > Biology > ARAP Path Interpolation.
Step-by-Step Overview
Step 1: Prepare Your Structures
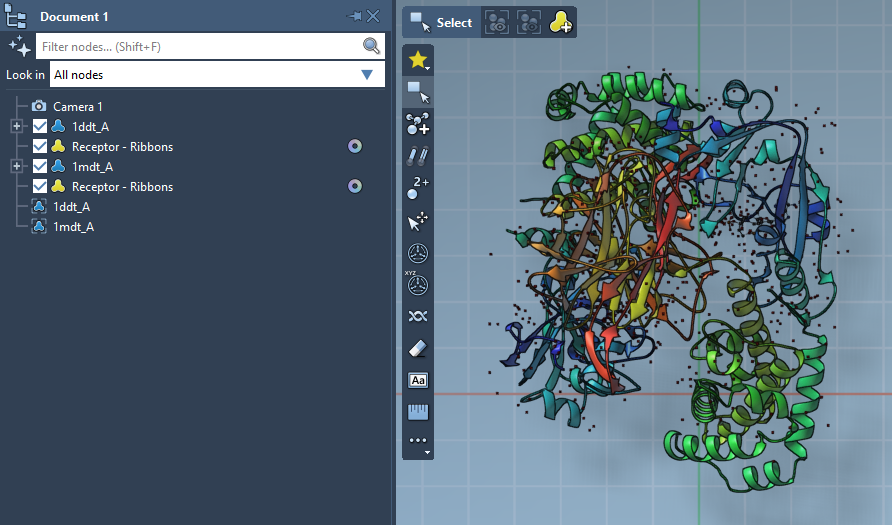
Fetch your initial and final structures directly from the PDB. For example, the conformations 1DDT and 1MDT represent two forms of the Diphtheria Toxin. You’ll likely want to keep only a single chain (e.g., chain A) and clean the structures with Home > Prepare.

Step 2: Define Conformations
Each structure needs to be marked as a distinct conformation. Select a cleaned structure, then go to Edit > Conformation and name it accordingly (e.g., “1DDT A” for the start and “1MDT A” for the goal).
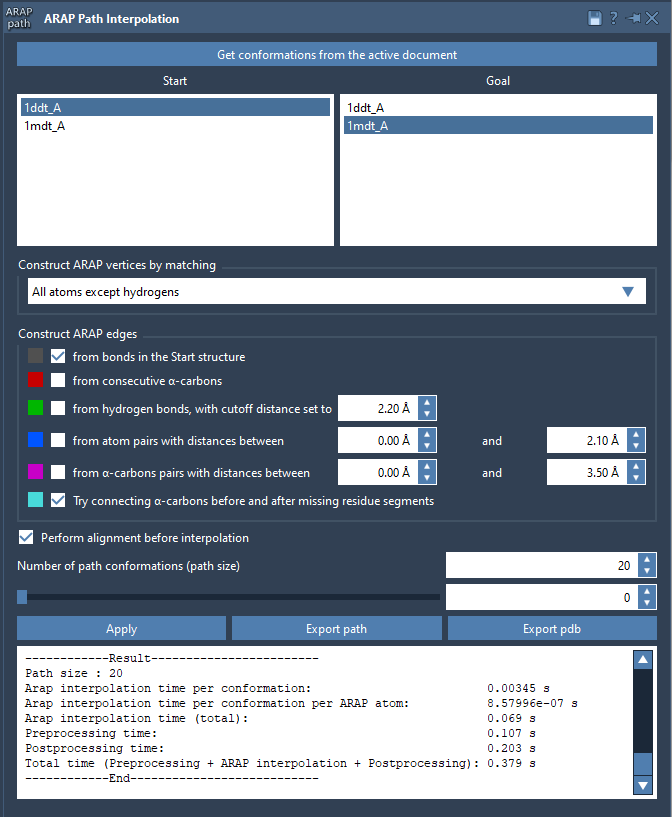
Step 3: Configure Interpolation Parameters
In the ARAP Path Interpolation app:
- Select your start and goal conformations using the provided button.
- Match atoms (commonly All except hydrogens) and check ARAP edge options such as from bonds in the start structure and connect alpha-carbons before and after missing residue segments.
- Enable alignment before interpolation and set how many conformations you’d like to generate (e.g., 20).
Step 4: Run and Visualize
Click Run, and SAMSON computes a trajectory between the two structures. You can scrub through frames using the app slider, view ARAP edges that show motion continuity, and analyze trajectory files or export to PDBs.
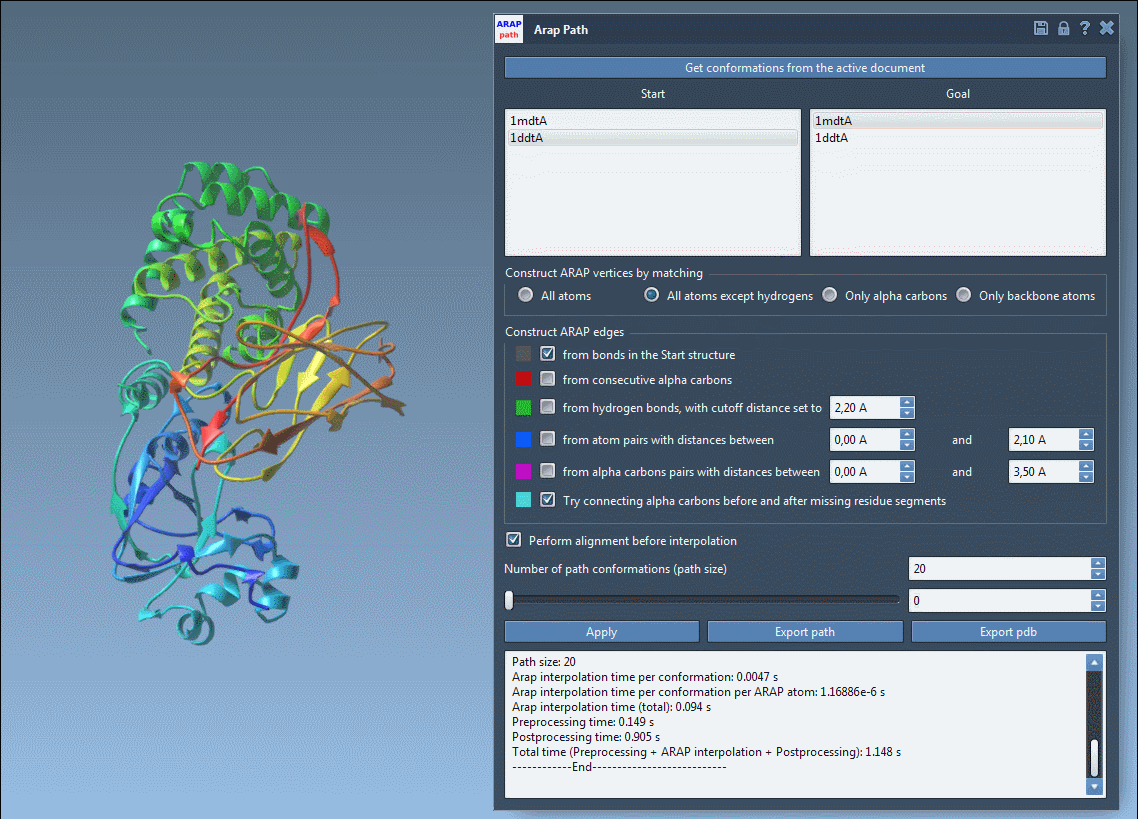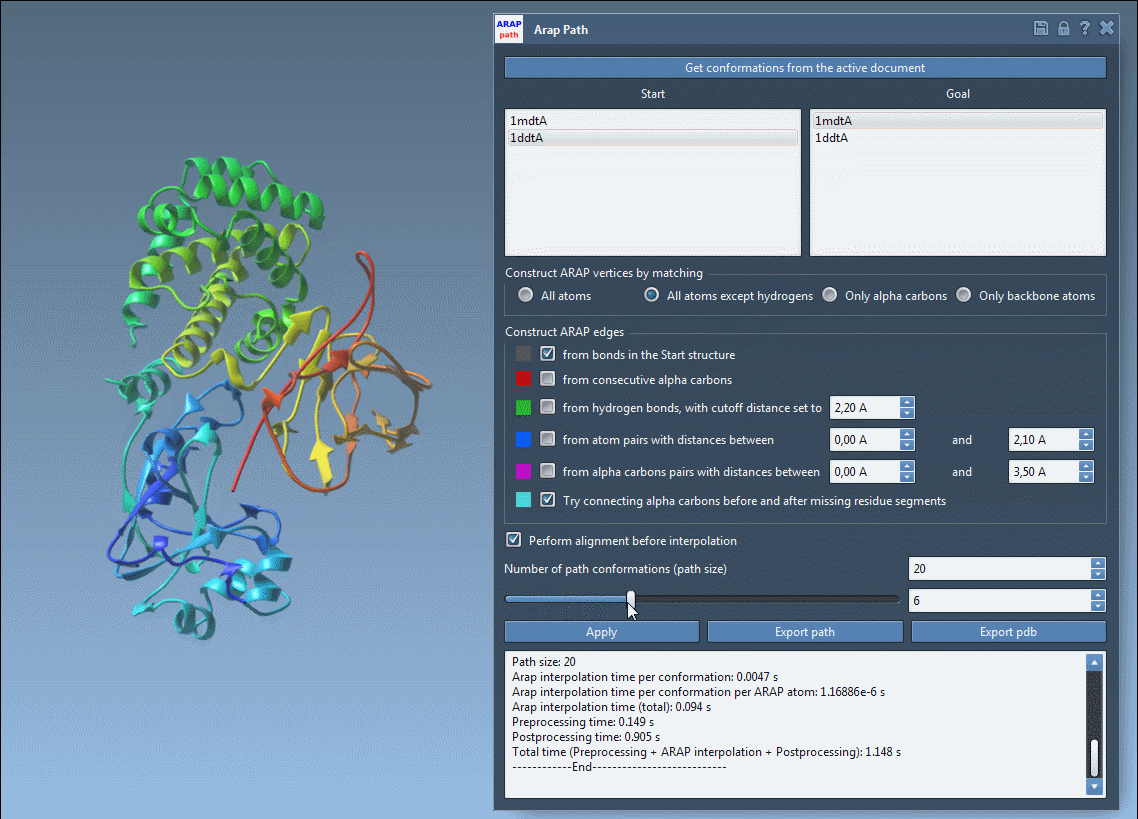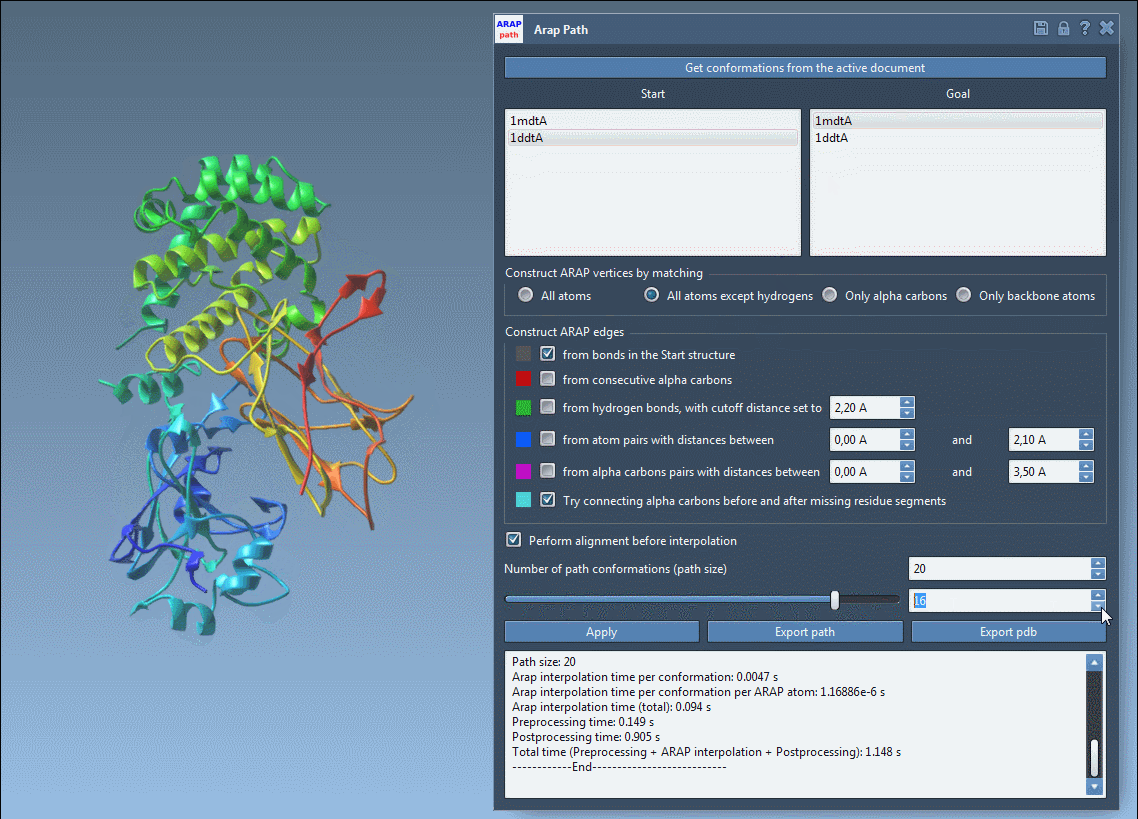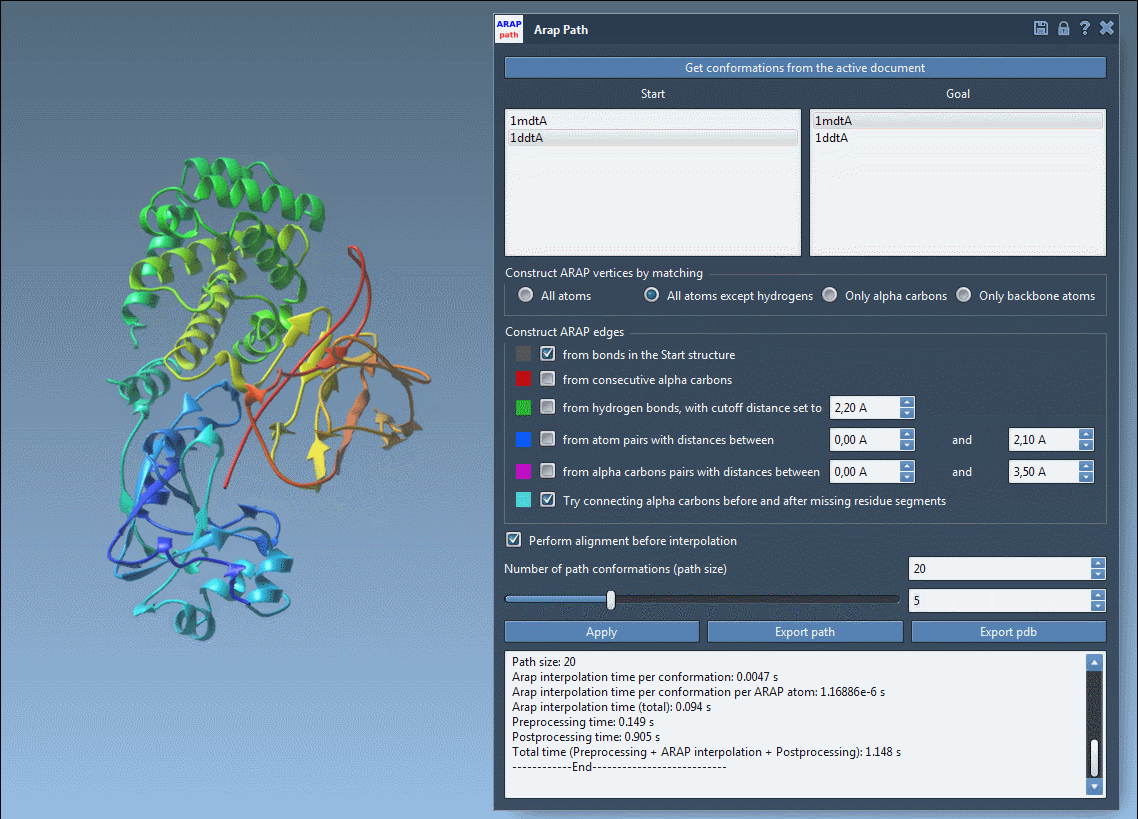
What It Enables
ARAP interpolation doesn’t require simulation parameters or complex force-fields. It quickly gives you a realistic molecular animation that can serve as:
- A visual hypothesis for domain movement
- An input for methods like P-NEB or umbrella sampling
- An educational illustration of conformational flexibility
For many molecular modelers, this may be the missing step between PDB discovery and simulation setup.
To learn more, visit the full tutorial page: Generating a Transition Path Between Protein Structures with ARAP Interpolation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





