When analyzing or designing protein structures, molecular modelers often encounter the challenge of working with incomplete biological assemblies. PDB files offer a rich source of structural data, but this data can be encoded in multiple ways — complicating the reconstruction of full assemblies or symmetric complexes.
If you’ve ever wondered whether to use CRYST1 or BIOMT records from a PDB file for generating symmetry mates, or if you didn’t know there was a difference at all, this post is for you.
Two Paths to Symmetry
In PDB files, protein symmetry information is typically encoded in one of two places:
- CRYST1: Describes symmetry based on the crystal lattice. These symmetries are derived from the repeating units found during X-ray crystallography and are useful when studying the crystal environment.
- BIOMT: Defines the biological assembly, indicating how the authors of the structure believe the functional multimer exists in vivo. This is often the symmetry that modelers want for simulations and design applications.
Models that rely solely on the asymmetric unit may be missing essential biological context. Being able to switch between these two representations on the fly can make a significant difference when building or analyzing quaternary structures.
CRYST1 vs. BIOMT in Practice
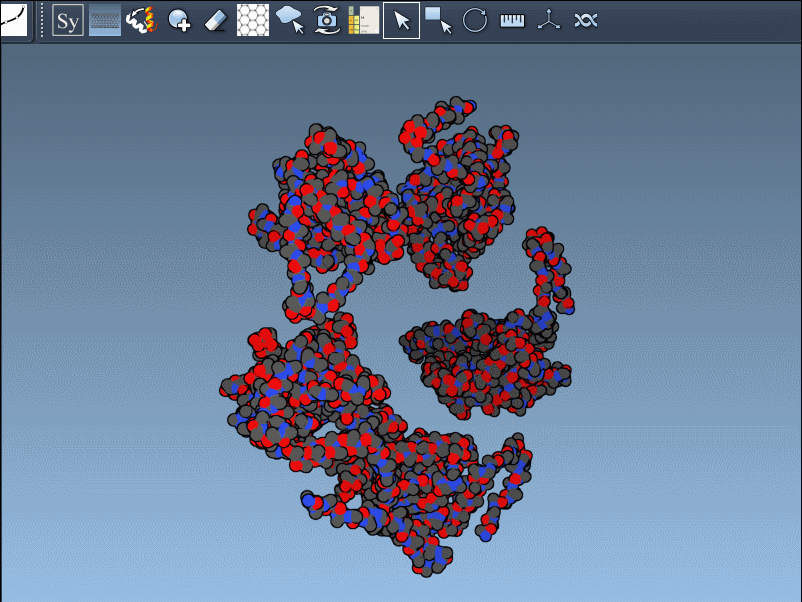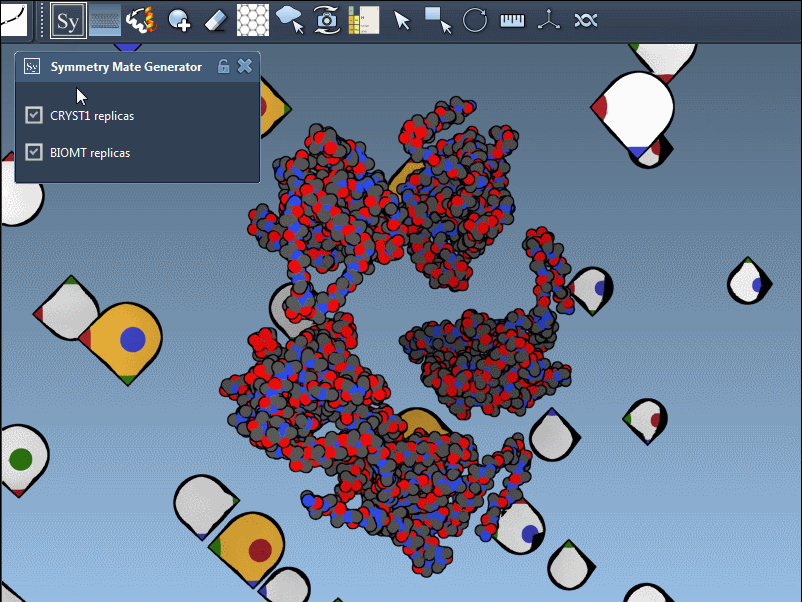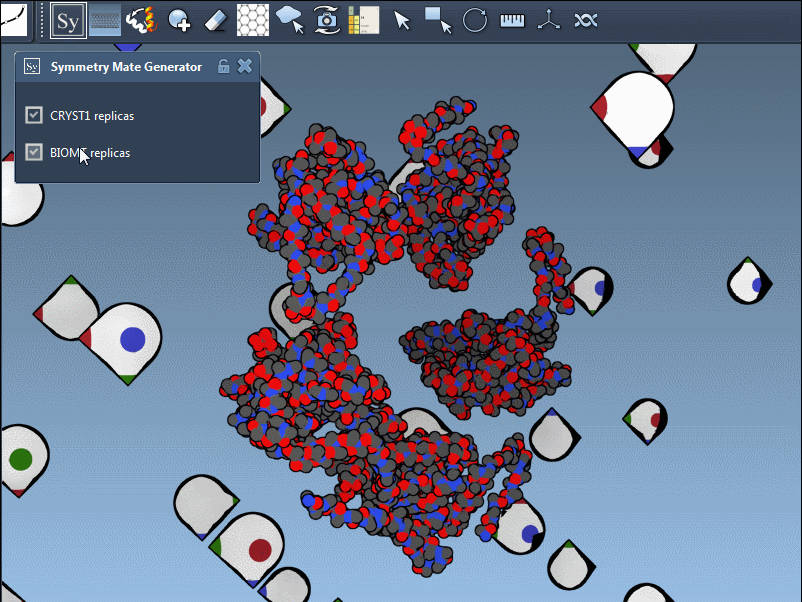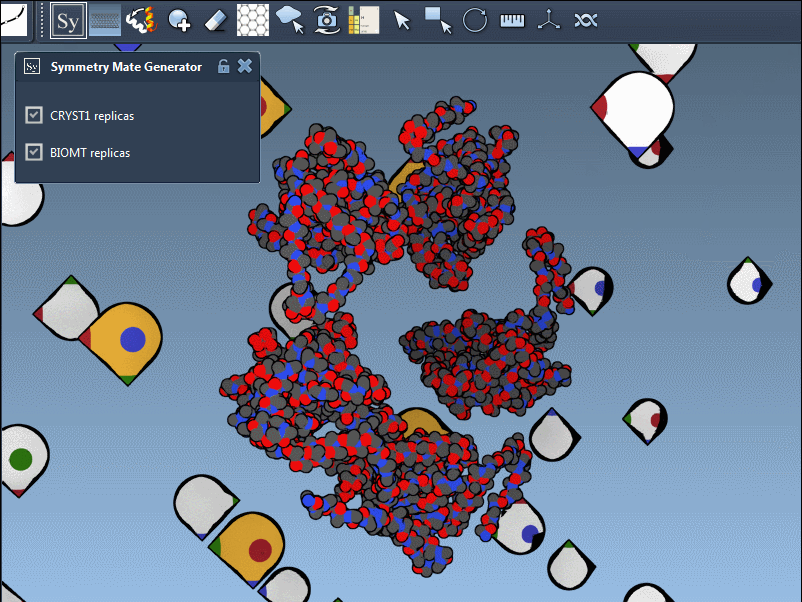
In SAMSON’s Symmetry Mate Editor, distinguishing between these symmetry types is made visually intuitive. When using the editor:
- White control nodes correspond to CRYST1-derived symmetry mates.
- Yellow control nodes indicate BIOMT-based symmetry mates.
This color coding helps ensure you’re always aware of the symmetry context you’re working within.
When to Use What
If your goal is to:
- Reconstruct the biologically relevant multimer — use BIOMT.
- Study crystal packing or lattice interactions — use CRYST1.
In many cases, exploring both can be valuable, especially when preparing datasets for simulation or machine learning experiments. Starting from the correct assembly avoids downstream structural artifacts and improves biological relevance.
Switching Between Them in SAMSON
Thanks to SAMSON’s integrated interface, toggling between CRYST1 and BIOMT symmetries is fluid. Simply activate the Symmetry Mate Editor, then select which symmetry you’d like to work with using the editor’s controls. You can preview replicas by hovering over nodes, and generate them with a click.
Here’s an example using PDB ID 1B5S, showing how the same protein structure differs depending on which symmetry is used:

Conclusion
The distinction between CRYST1 and BIOMT isn’t just technical jargon; it can shape the outcome of your structural analysis or design project. By understanding the origin and intent of each symmetry type and using a tool like SAMSON that makes them accessible and explorable, you can streamline your modeling workflows and improve the accuracy of your structural interpretations.
To learn more, visit the full documentation page here.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





