Tracking the movement of specific molecular components across a simulation is a common task in molecular modeling and simulations. One particularly useful metric is the motion of the center of mass (COM) of a group of atoms — for instance, a ligand moving through a protein during an unbinding event.
However, visualizing such motion in a meaningful, interpretable way can be time-consuming or require custom scripts. That’s where the Pathlines extension in SAMSON comes in. It provides a way to graphically plot the COM trajectory of a selected group of atoms along one or more paths built from other simulations or methods like the Ligand Path Finder.
Why Track Center-of-Mass Motion?
In complex molecular systems, it’s often more insightful to look at the trajectory of an entire molecular group’s COM — especially when analyzing:
- Ligand unbinding or rebinding events
- Protein domain reorganizations or transitions
- Collective atomic behavior over a path
COM pathlines reduce noisy atomic-level detail into smooth representations that are easier to interpret, align, and compare.
How to Visualize COM Motion with Pathlines
Let’s walk through the workflow using a sample system included in the Pathlines tutorial, step by step.
Step 1 – Load the Sample System
To get started quickly, download the sample system from SAMSON Connect which includes the lactose permease (1PV7) with its ligand TDG and predefined unbinding paths.

Step 2 – Select Atoms and Paths
In the Document view, select the ligand atoms (e.g. TDG) and one or more paths that represent different unbinding trajectories. If you select nothing, SAMSON will default to all atoms and all paths.

Step 3 – Create the Pathline Visual Model
Now go to Visualization > Visual model > More… or use the shortcut Ctrl/Cmd + Shift + V. Choose Pathline of the center of mass in the dialog, and click OK.
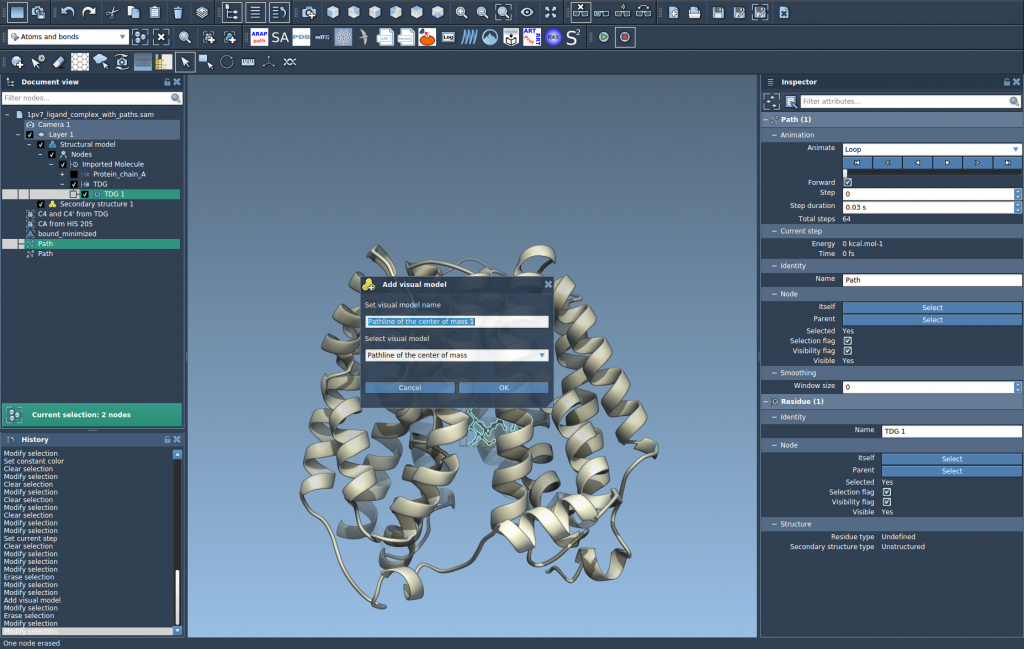
This generates a pathline showing how the COM of the selected atoms (like your ligand) moves through the selected paths.
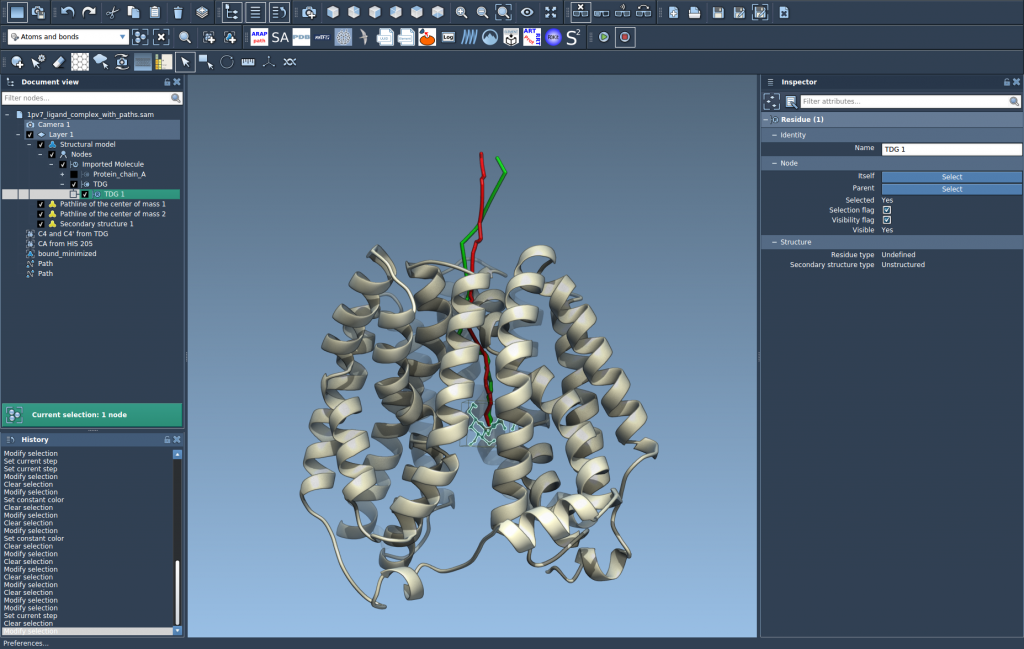
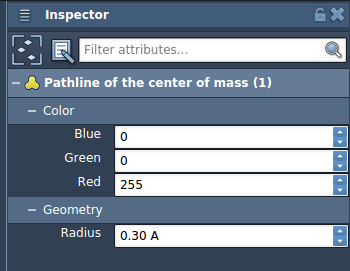
Step 4 – Customize Your Visualization
Use the Inspector (Ctrl/Cmd + 2) to customize the pathline. You can adjust its color, thickness, and style to make it easier to read or highlight key differences across simulations.
When Are Pathlines Most Useful?
Pathlines are especially useful when analyzing or presenting your results, or trying to make sense of ensemble data created by automatic path generation algorithms. Use cases include:
- Displaying ligand unbinding/rebinding routes
- Comparing multiple possible paths in a reaction coordinate
- Studying domain motions in protein complexes
The visual clarity provided by COM pathlines makes this tool valuable for inspections, presentations, and publications.
To learn more, check the full Pathlines documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





