For molecular modelers investigating viral infection mechanisms or working on therapeutic design, understanding protein conformational transitions is a frequent and sometimes challenging task. Particularly in virology, capturing how viral proteins change shape to interact with host receptors can help drive hypotheses on drug binding, vaccine effectiveness, or antibody evasion.
One recent case that has sparked interest worldwide is the SARS-CoV-2 spike protein, responsible for helping the virus enter human cells. But this doesn’t happen with the spike just sitting still — it undergoes a noticeable mechanical shift from a closed (down) state to an open (up) state, which exposes the receptor-binding domain (RBD). Modeling and animating such conformational changes offers insights into how binding occurs and illustrates targets for neutralizing antibodies.
Challenges for Molecular Modelers
If you’ve ever tried modeling the transition between two structural states of a protein that differ in small but significant ways — say, in residue numbering, sugar placement, or bond connectivity — you know how tricky this can be. Interpolating between these states often requires more than just morphing geometry: it needs chemistry-aware paths that respect changes in structure while avoiding unrealistic intermediates.
How This Spike Motion Was Modeled
Using SAMSON, a team has computed and animated the structural transition of the SARS-CoV-2 spike protein from PDB structures 6VYB (open) and 6VXX (closed). Here’s a summary of their approach:
- Hydrogens and bond orders were standardized using Python scripting to make the molecules compatible with minimization and path generation tools.
- Energy minimization steps helped stabilize intermediate states.
- The ARAP Interpolation Path module created a physically plausible pathway between the open and closed states in seconds. It’s based on the As-Rigid-As-Possible deformation model, making it useful even for large biological assemblies.
- The path was further refined using the Parallel Nudged Elastic Band (P-NEB) module, improving theoretical accuracy by considering force-based energy gradients along the conformational transition.
These steps resulted in a smooth, science-based animation of the spike opening.
See It in Motion
The outcome is not just numerical: the motion is visualized in several angles, making it accessible for presentations, exploration, or further development:
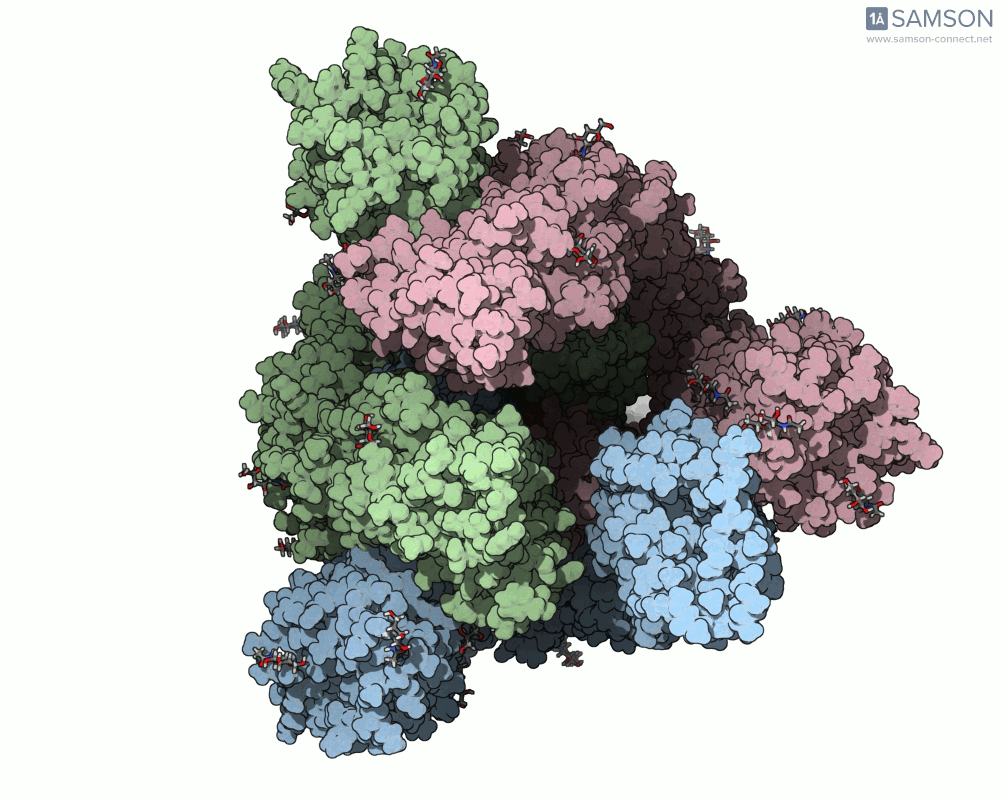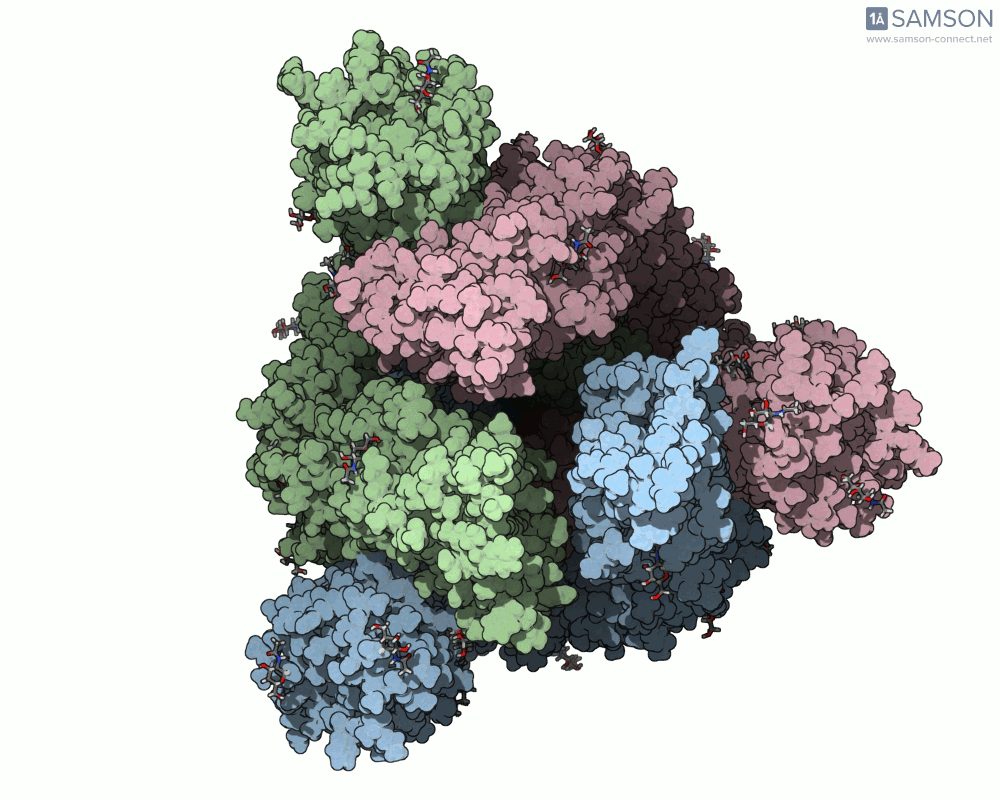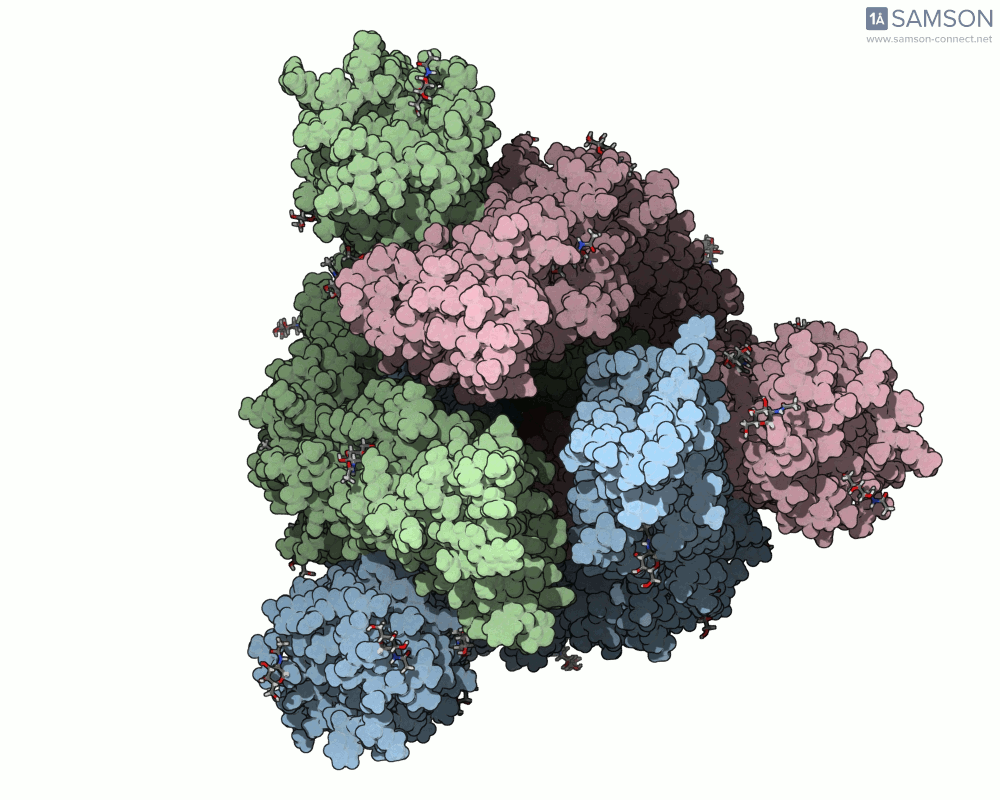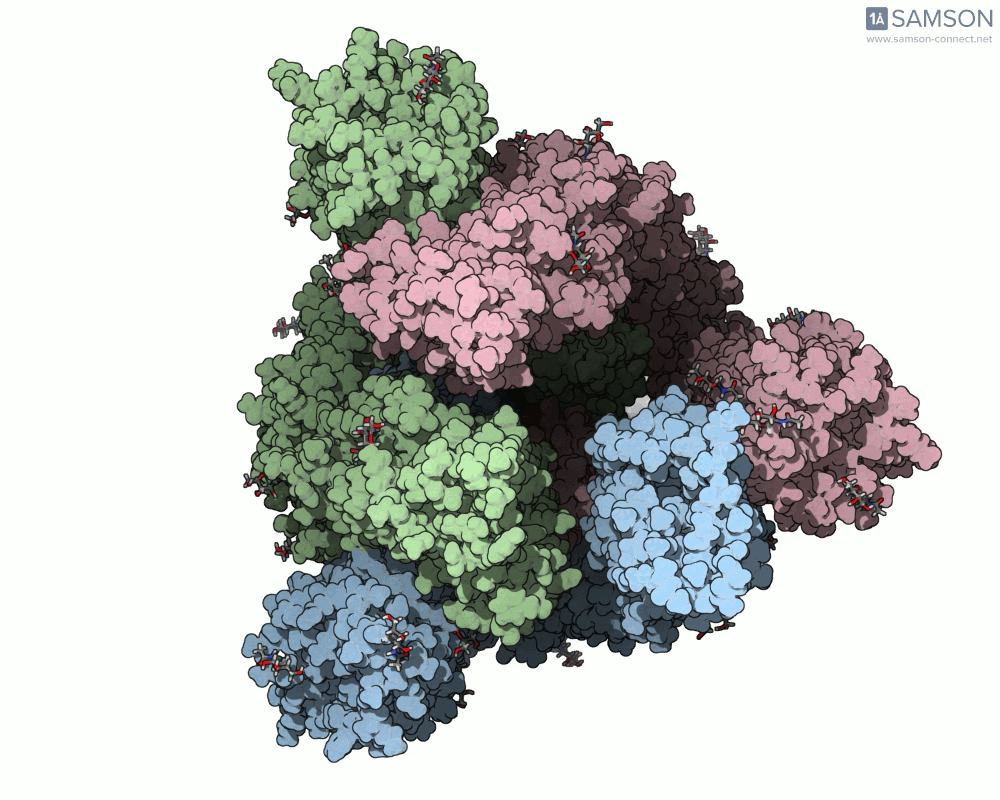
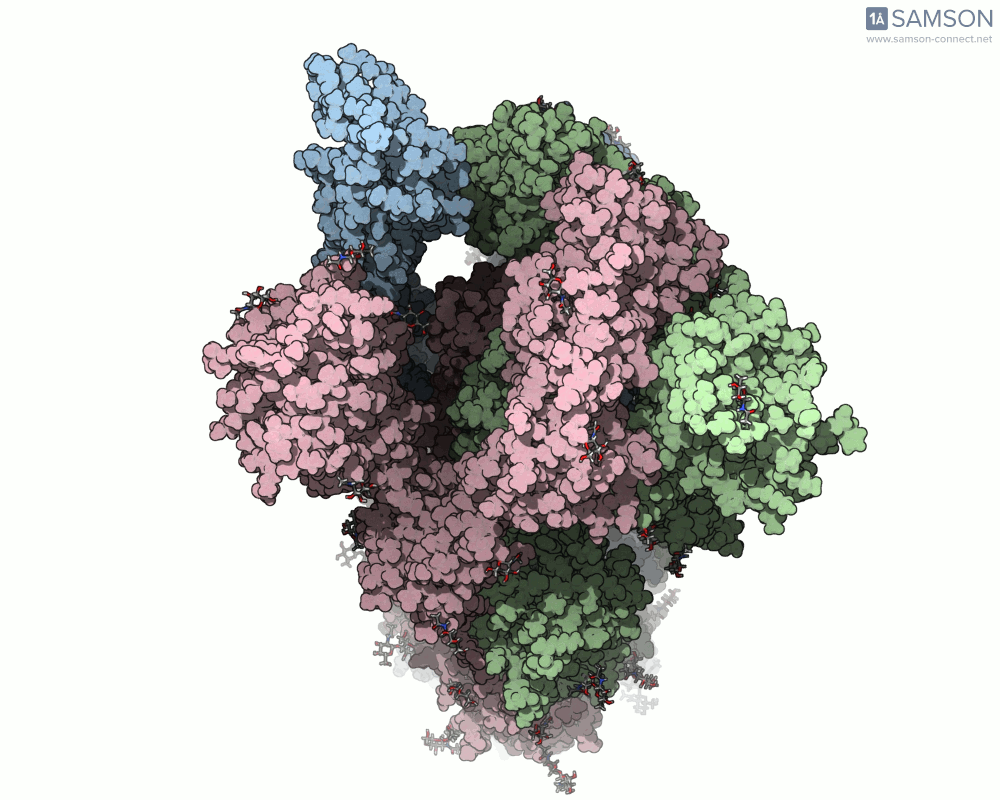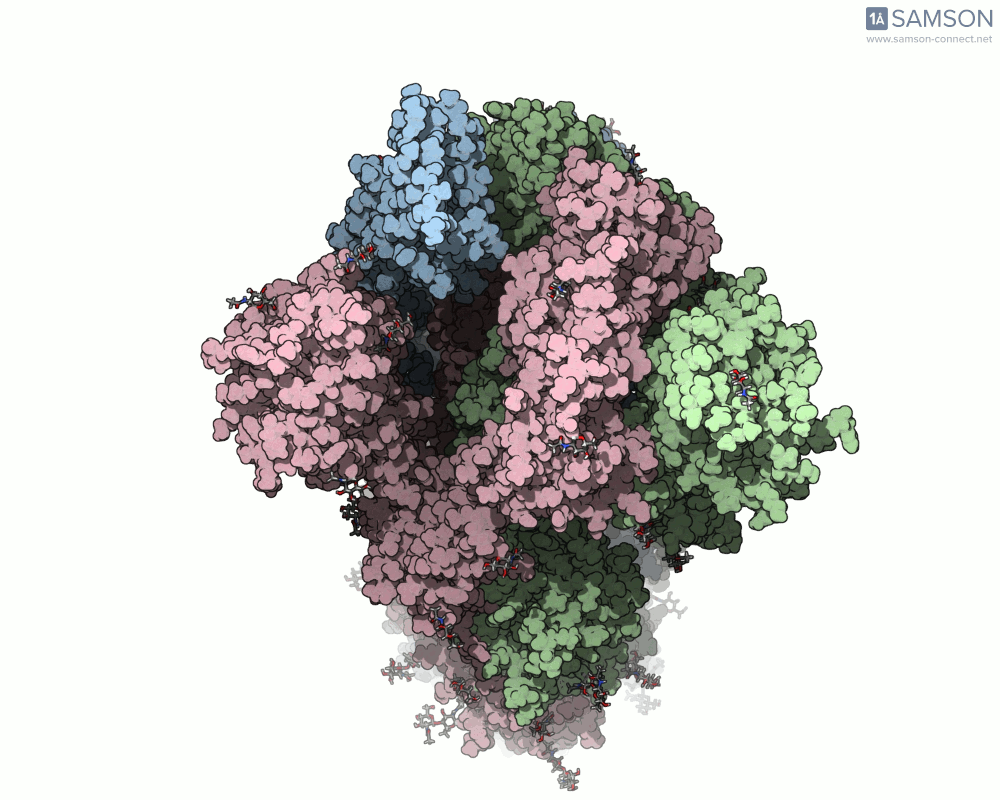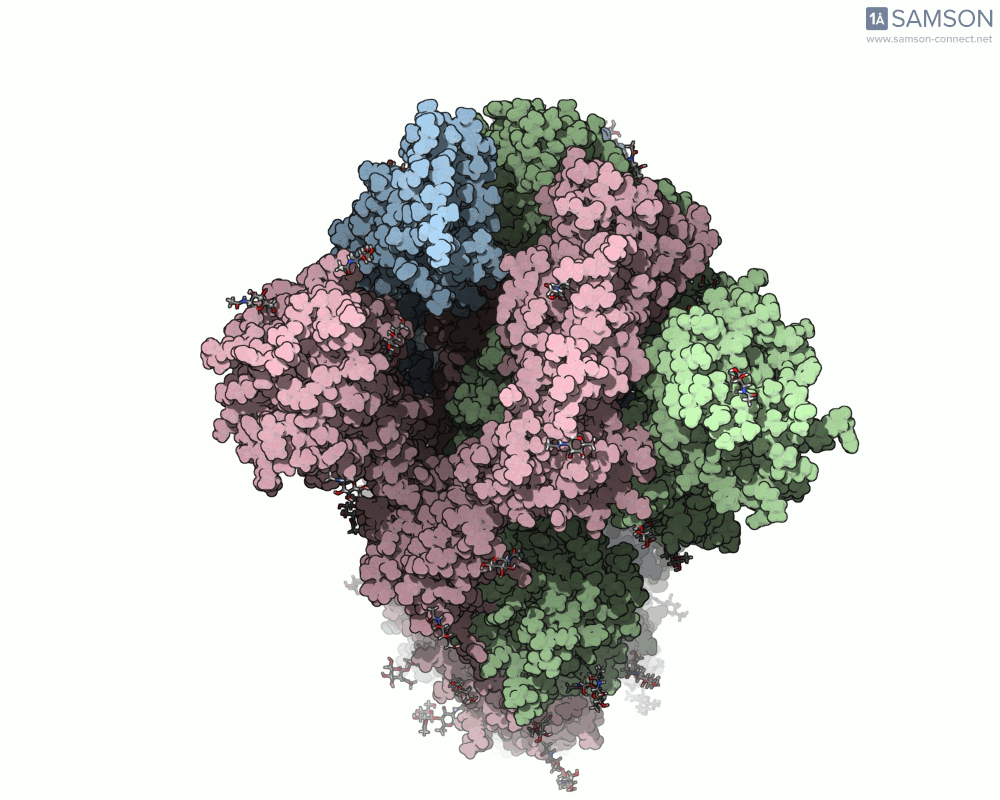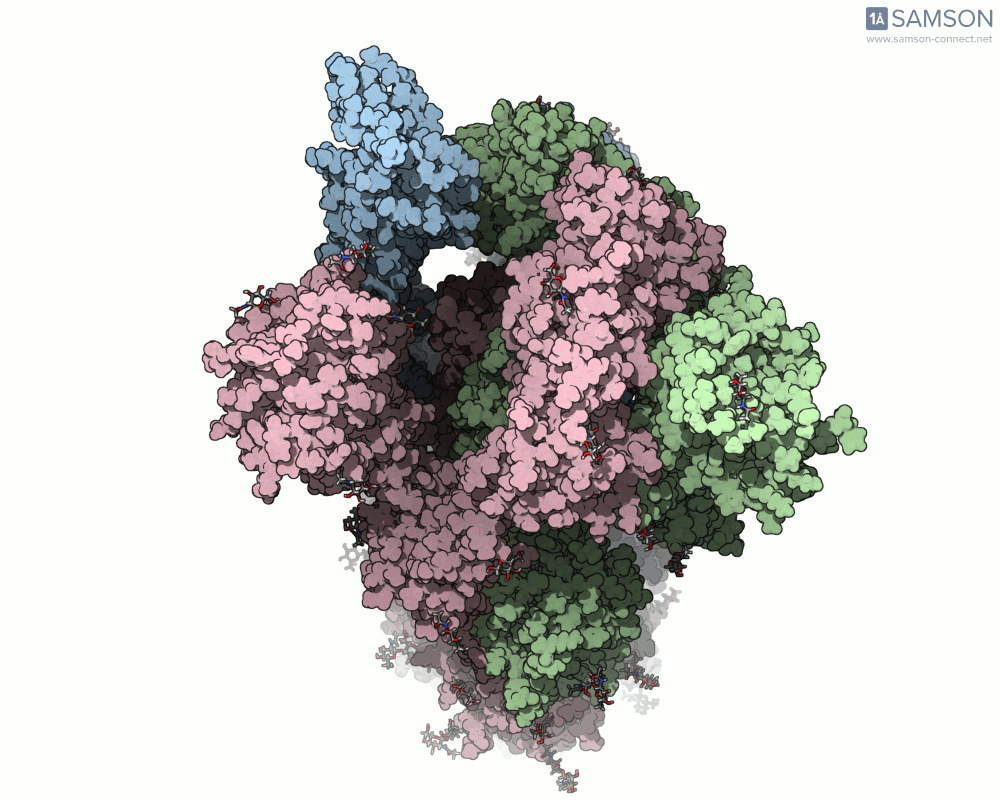
The SAMSON file includes the computed trajectory and both ARAP and P-NEB paths, ready for further exploration or refinement, and you can download it directly from the original article — no need to recreate the wheel.
Who Can Benefit from This?
Whether you’re studying viral mechanisms, designing therapeutic interventions, or teaching molecular biology, this ready-made example is both illustrative and a practical starting point. It highlights how conformational change modeling is more efficient — and insightful — when robotics-inspired methods are applied to molecular structures.
To explore the complete documentation and download the spike conformational trajectories, visit the full tutorial here.
SAMSON and all SAMSON Extensions are free for non-commercial use. Download SAMSON at https://www.samson-connect.net.





