One of the most common difficulties faced by molecular modelers is how to generate meaningful animations of conformational transitions between known protein states—especially when those states do not perfectly match in residue count or structure. Such animations are invaluable for both communication and early-stage hypothesis generation, particularly when studying biologically crucial systems like viral proteins. In this blog post, we’ll take a deep look into how the spike protein of SARS-CoV-2 transitions from its closed state to its open, receptor-binding state, and how you can reproduce this motion yourself using SAMSON.
Understanding the spatial dynamics of the SARS-CoV-2 spike is critical: only in its open state can it recognize and bind to the ACE2 receptor, triggering viral entry into human cells. To help visualize this transformation, the SAMSON team generated detailed animations using trajectory interpolation methods between experimentally available spike conformations.
The Biological Context
The spike protein is made up of three identical S proteins forming a trimer with C3 symmetry. It undergoes a conformational change from a closed state (PDB 6VXX) to an open state (PDB 6VYB). In the open configuration, one domain rises up, exposing the receptor-binding domain (RBD) that connects with ACE2 on the host cell surface.
The Molecular Animation Pipeline
The challenge? These structures differ in their residue counts and sugar modifications. Here’s how SAMSON tackled it:
- Pre-processing structures: Bond orders of sugars were standardized using a Python script to enable hydrogen addition, followed by geometry minimization.
- ARAP path computation: Using the ARAP Interpolation Path module, an initial transition path between the open and closed states was generated. This takes under 30 seconds on a typical laptop.
- Residue mismatch workaround: Since 6VYB and 6VXX have different residue counts, a hybrid minimized intermediate matching the open structure residues was created for consistent interpolation.
- P-NEB refinement: To improve the physical plausibility of the motion, the P-NEB module was used to relax the path. Total time: ~15 minutes on a laptop.
The result? A smooth, data-driven animation of the spike transitioning through intermediate states. These can be exported as trajectories (PDB or SAMSON format) for further analysis or visualization.
Watching the Spike in Action
The animation below shows the spike opening from a closed state (all RBDs down) to an open state (one RBD up):
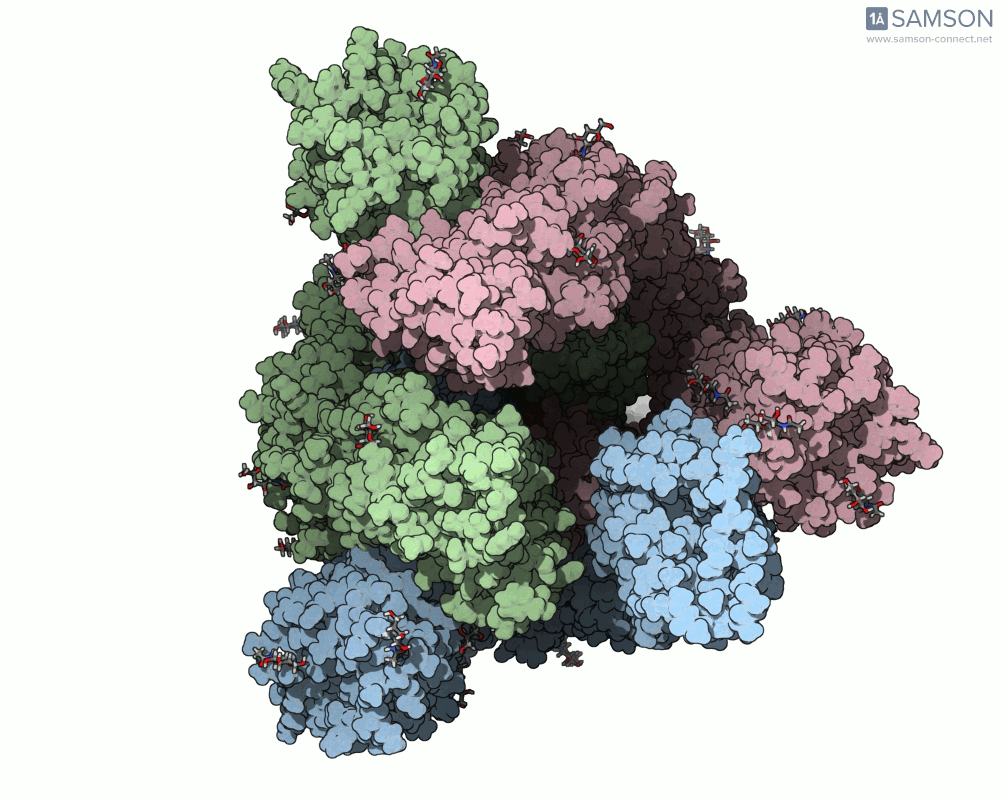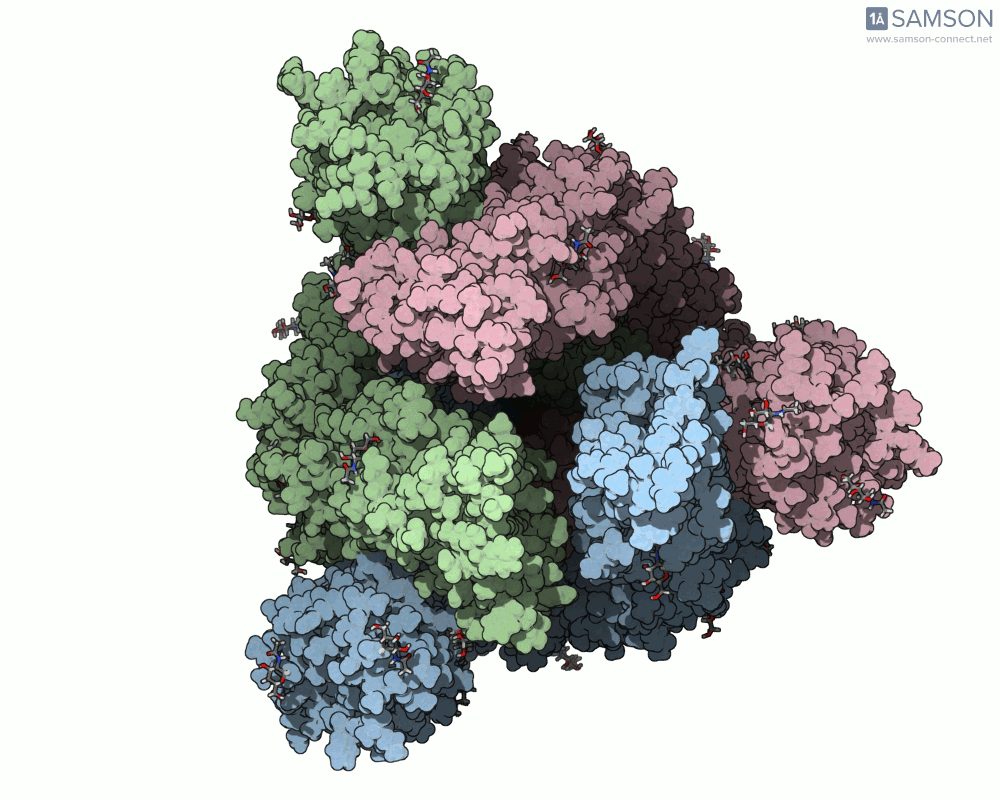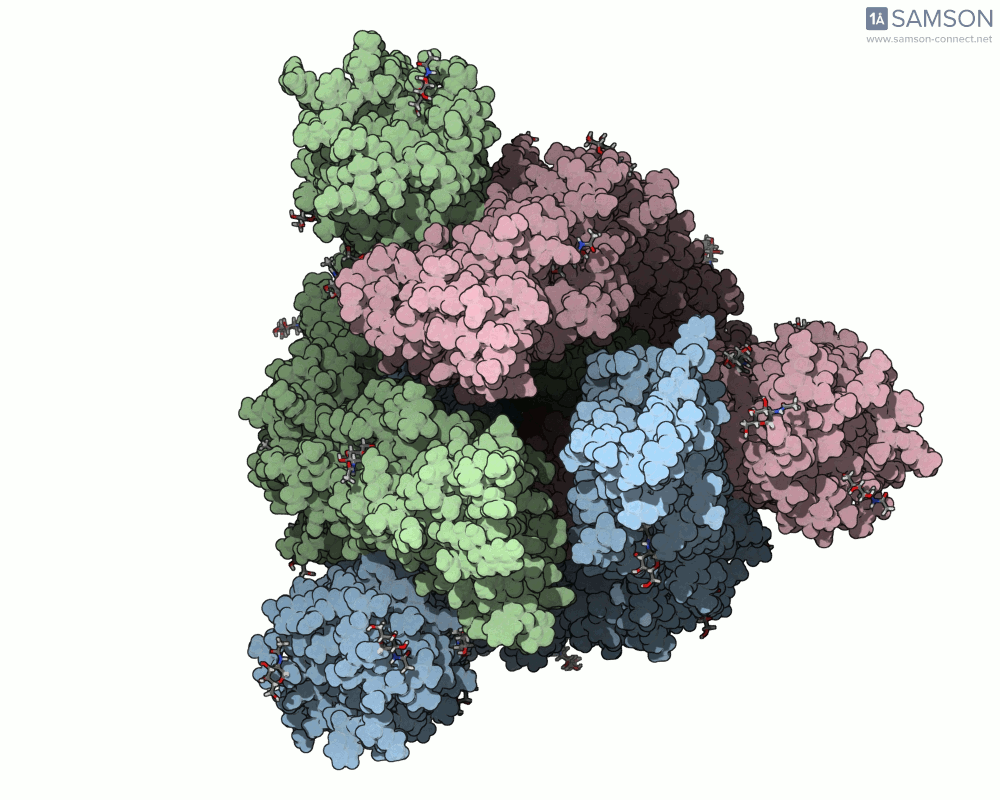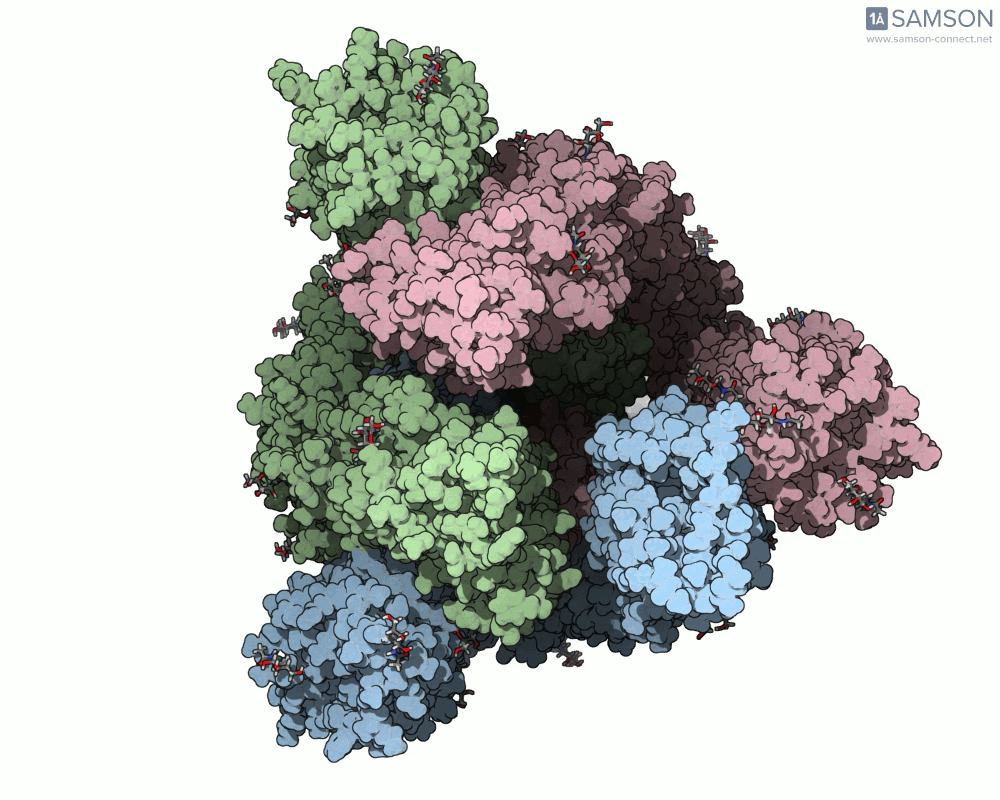
Such molecular animations are not only visually compelling but can aid in understanding target accessibility, antibody binding potential, and help with rational design of inhibitors or vaccines.
Want to Try This Yourself?
You can download the computed trajectories directly from the SAMSON documentation, which also includes all scripts and modules used. The ARAP and P-NEB modules are freely available for anyone studying COVID-19 or related systems.
If you work with molecular animations and often face challenges connecting two states of a molecule in a biologically meaningful way—especially when experimental data is incomplete or structurally inconsistent—then this tutorial can offer a concrete and practical solution.
To learn more, visit the full SARS-CoV-2 spike motion tutorial on SAMSON documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON here.





