One common challenge in molecular modeling is visualizing conformational changes between known states of a complex biomolecule—especially when those changes reveal key biological functions, such as viral infection mechanisms. If you’ve ever worked with large protein structures that undergo dramatic conformational transitions, you’ve probably faced the question: How can I create a physically meaningful, visual model of this movement?
In this post, we explore how the SAMSON molecular design platform can help modelers generate and visualize the transition trajectory of the SARS-CoV-2 spike protein from its closed state to its open, receptor-binding state. This is particularly relevant for those modeling the effects of neutralizing antibodies or studying receptor binding mechanisms.
What makes this trajectory special?
The spike protein of SARS-CoV-2 is the entry key to human cells. Its opening motion allows binding to ACE2 receptors, a process critical for infection. Modeling this movement helps visualize how interventions like therapeutic antibodies block infection, and serves as a teaching tool or the basis for simulation experiments.
To do this, the SAMSON team used two reference structures:
They computed an interpolated pathway using SAMSON’s ARAP (As-Rigid-As-Possible) module, and refined it using the P-NEB (Parallel Nudged Elastic Band) module. The result is a smooth, visually intuitive trajectory of the spike opening motion.
Watch the spike in motion
Here are animation frames showing the trajectory from different views:
- Side view:
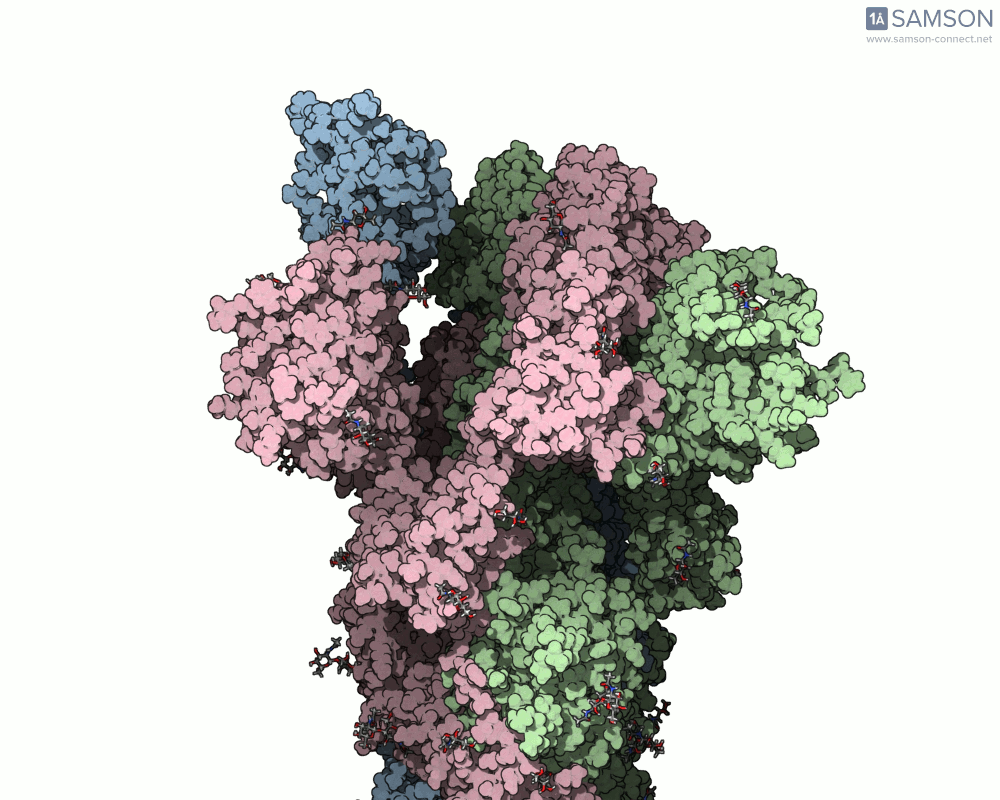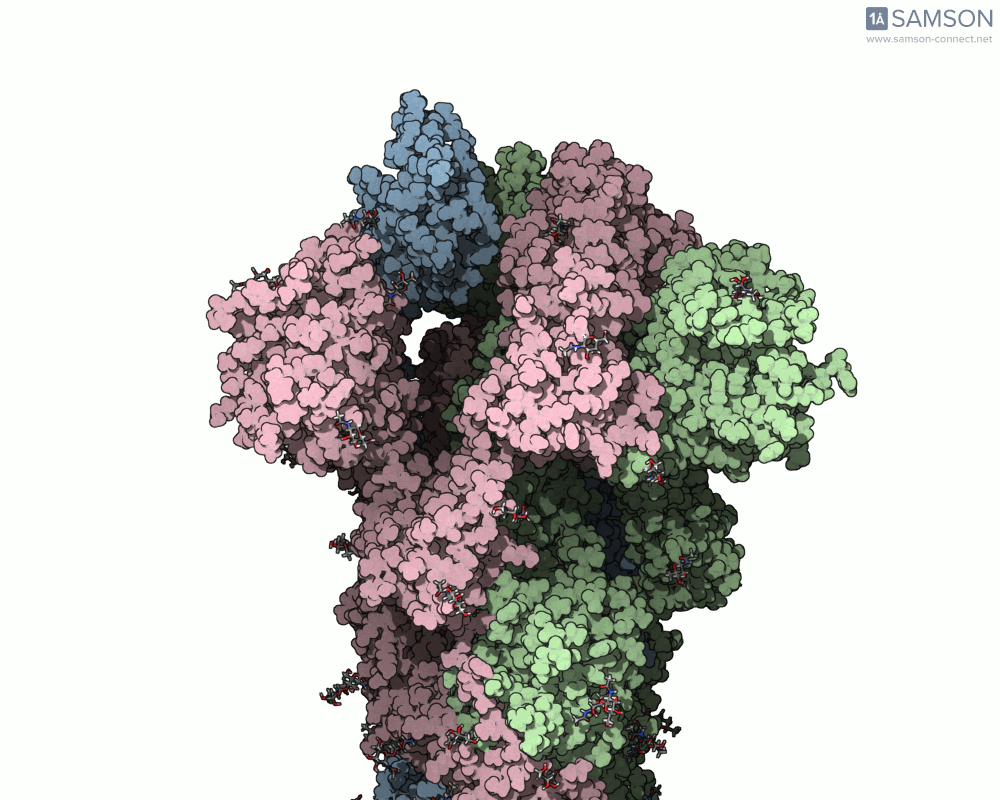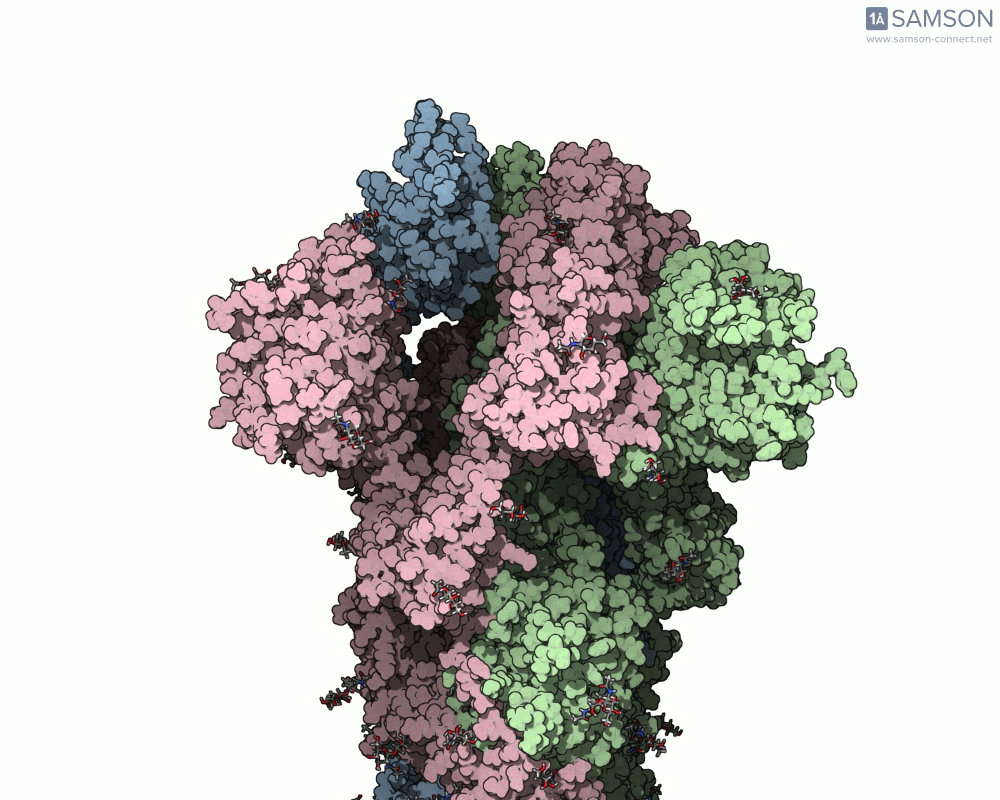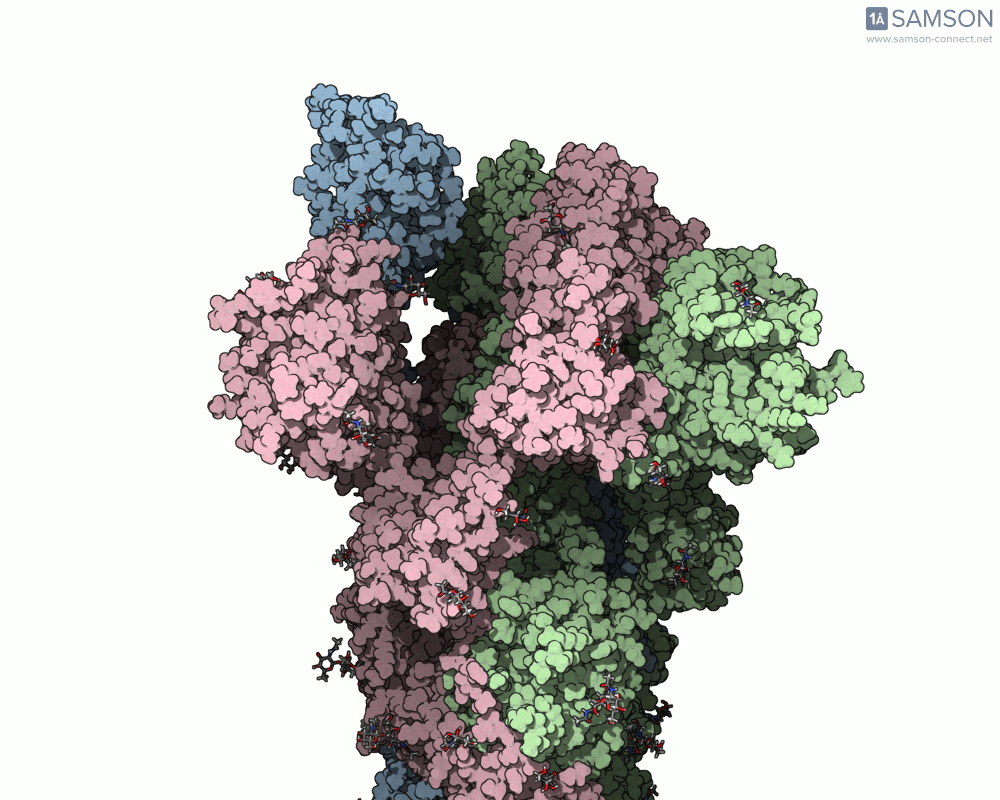
- Angled view:
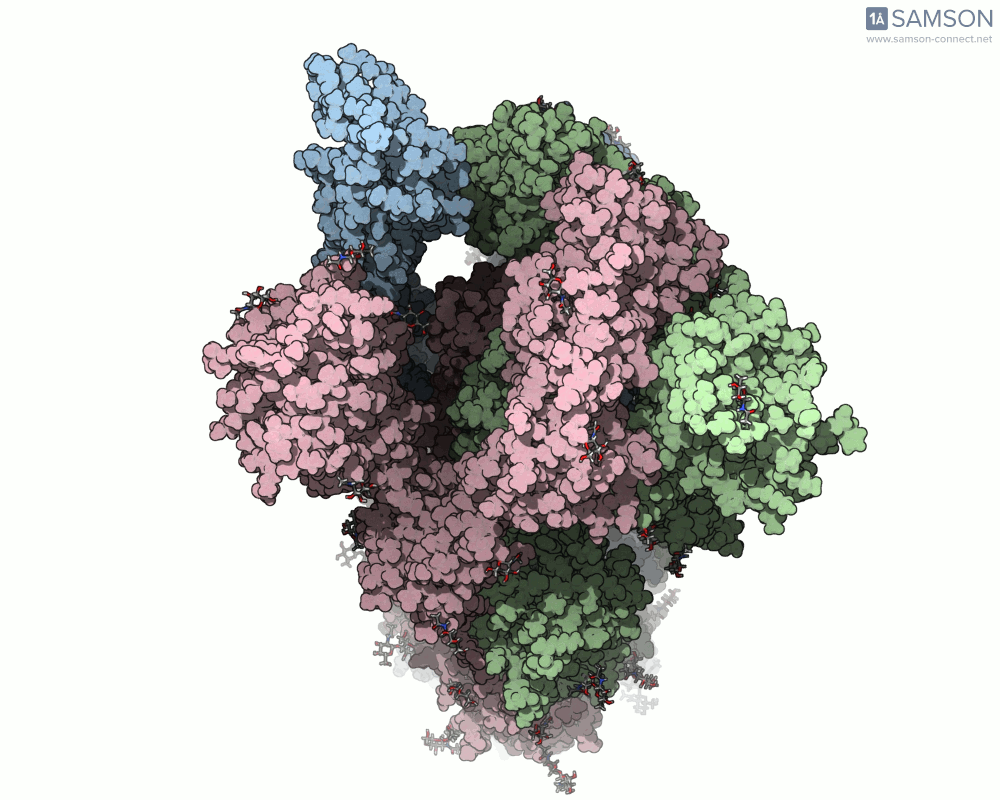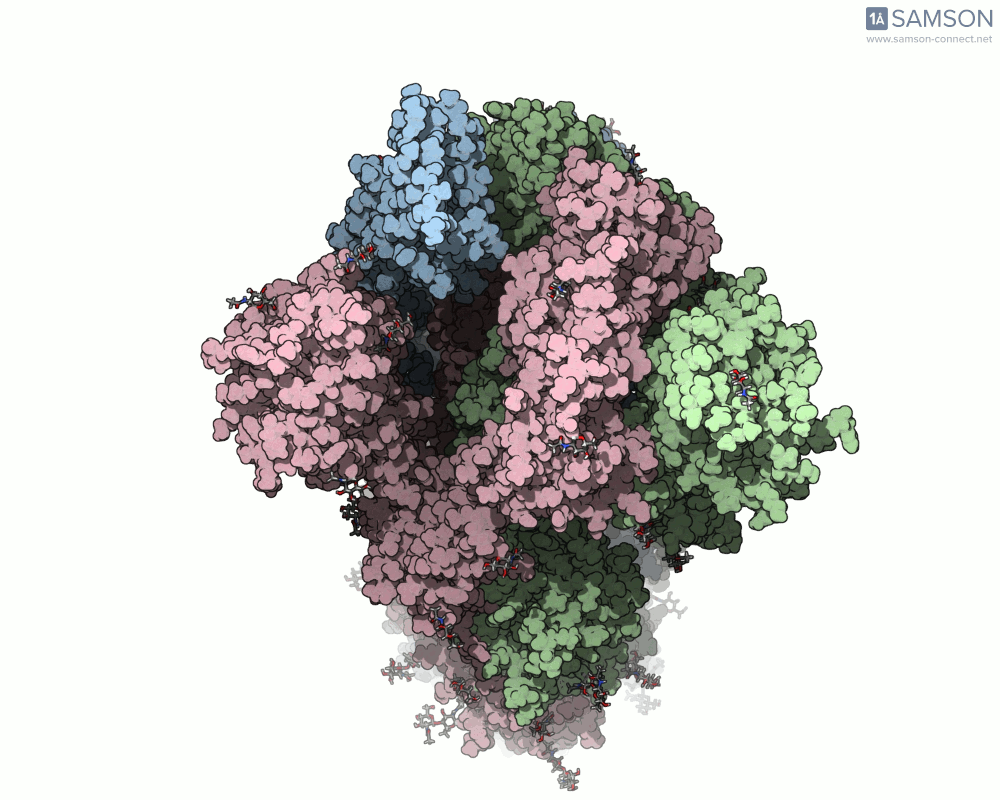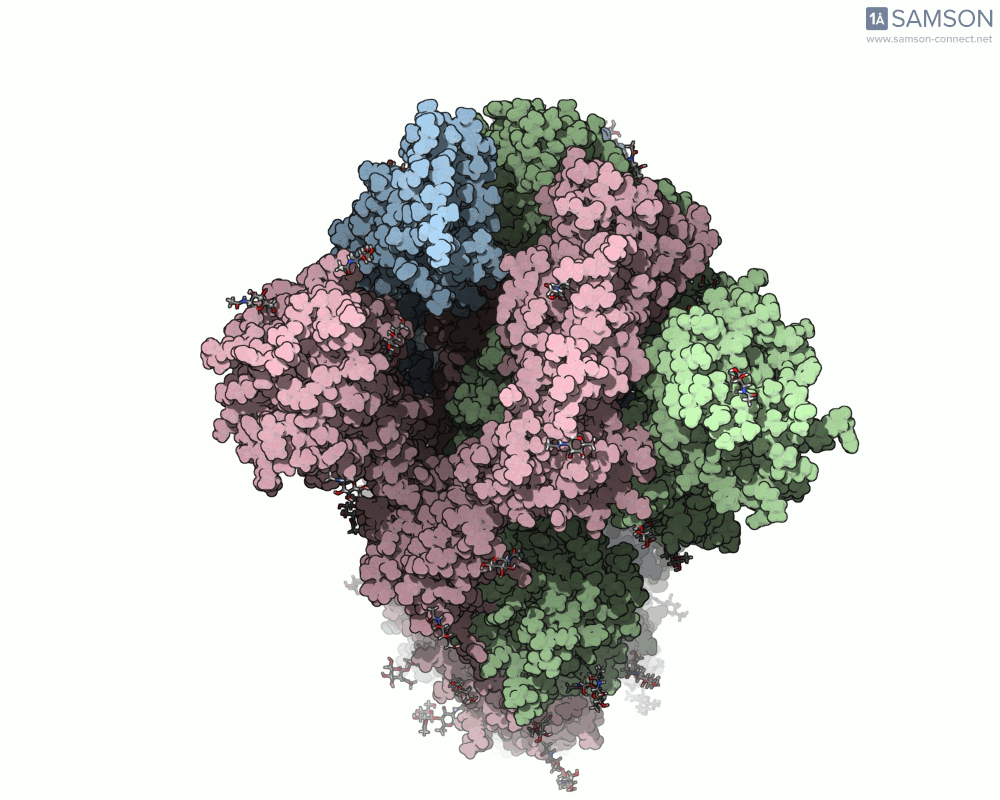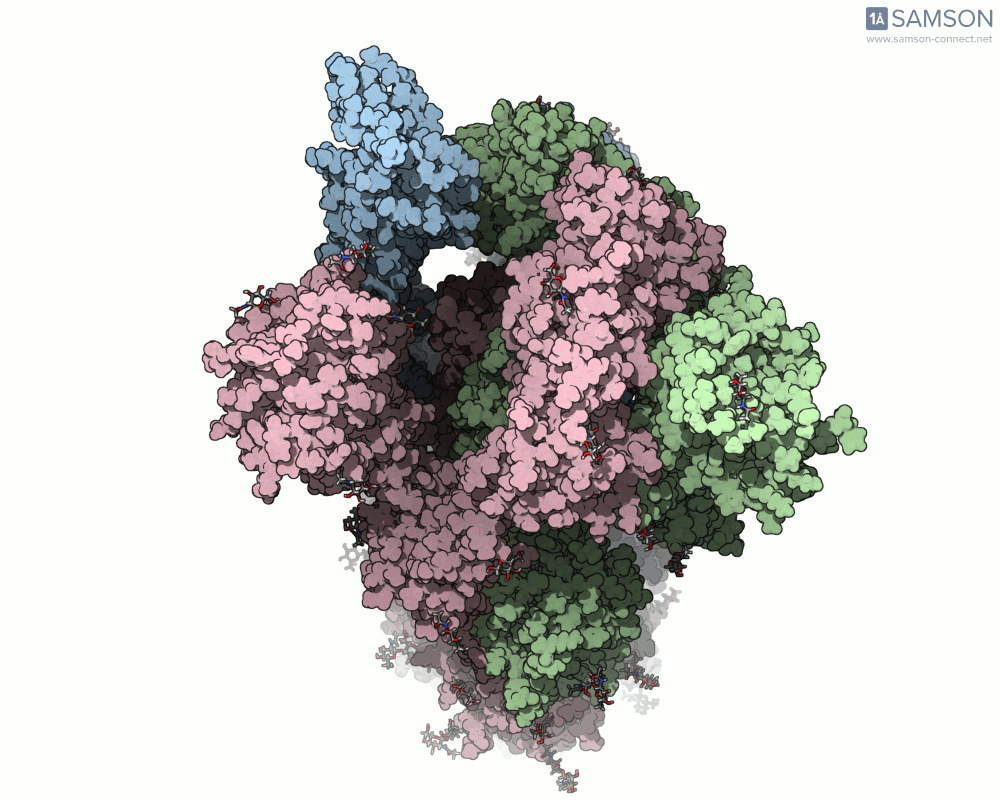
- Top view:
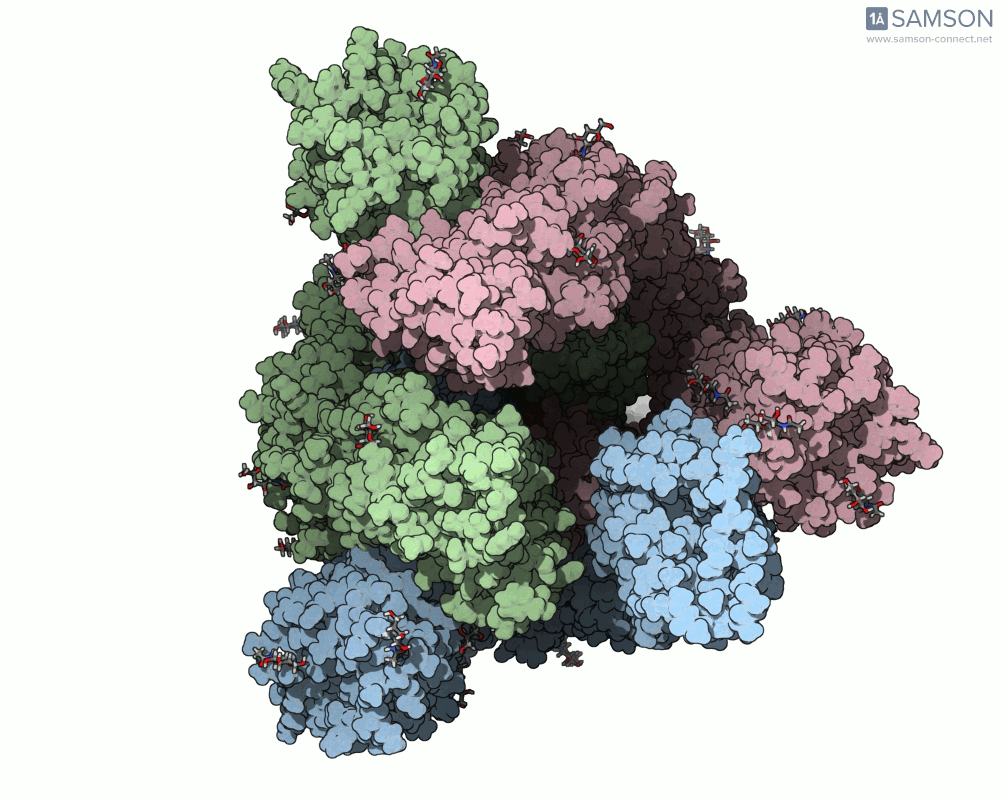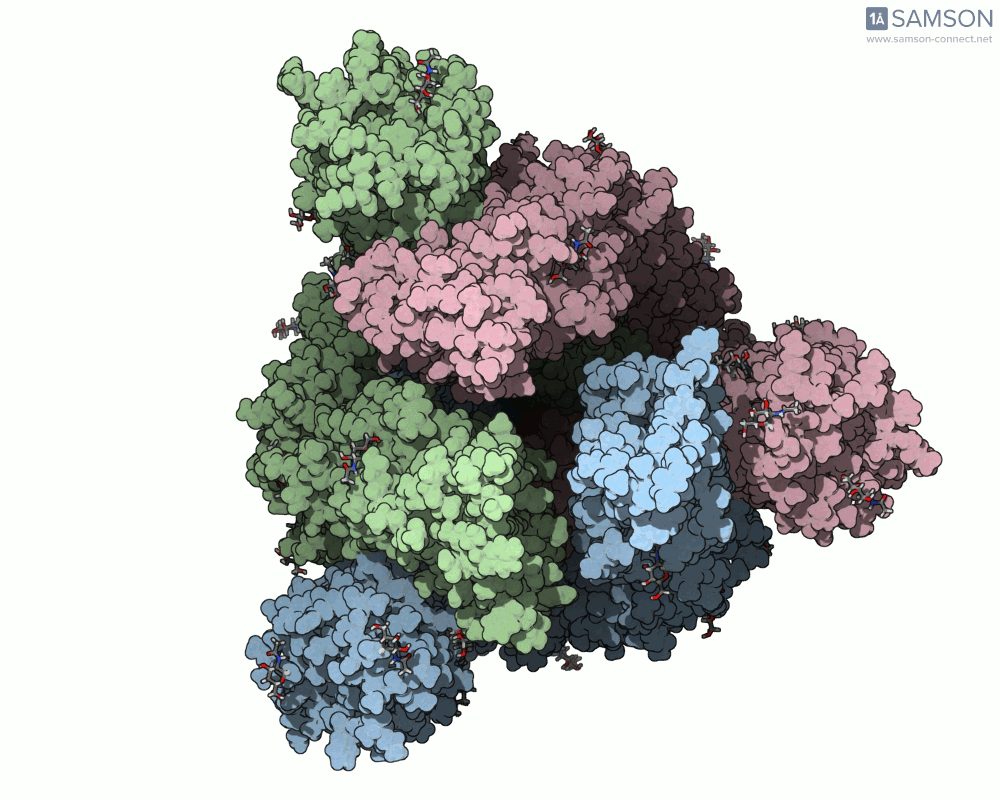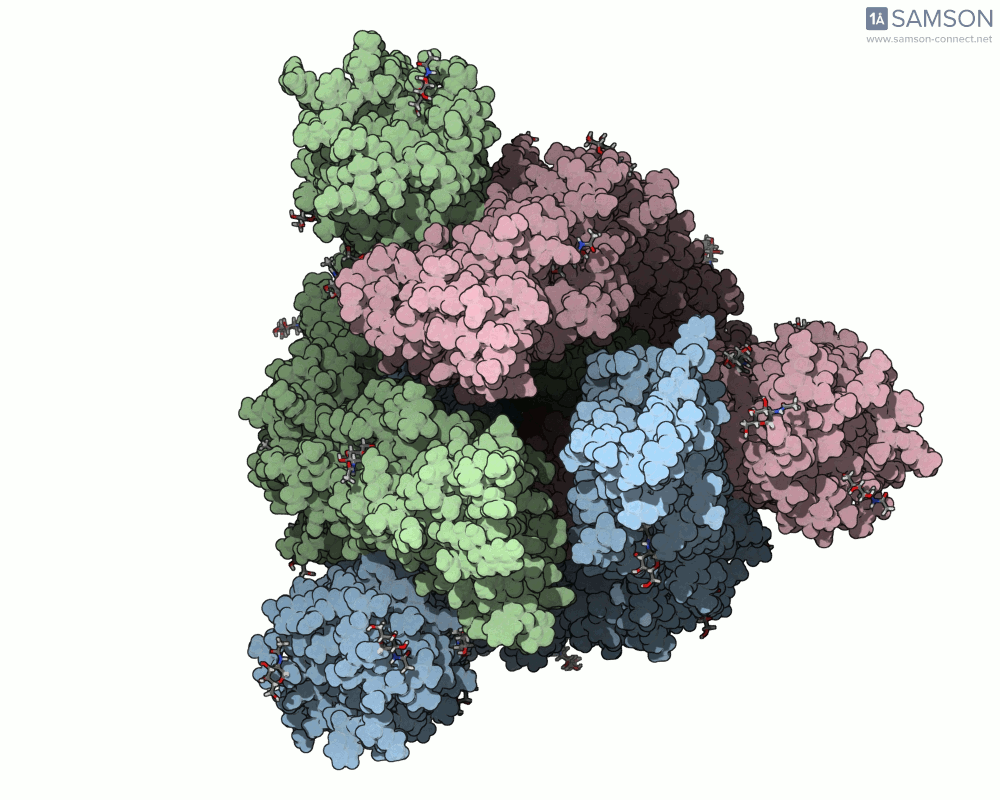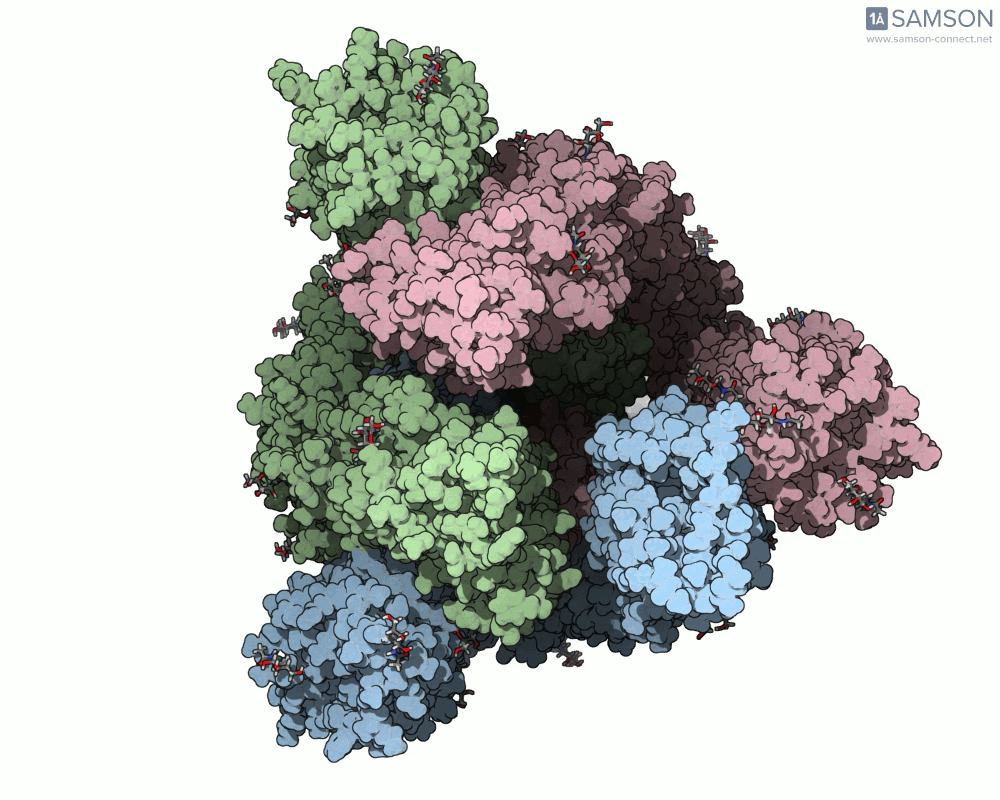
These visualizations help demystify the functional mechanics of a critical viral protein. They’re particularly valuable for communicating structural dynamics in academic and research settings.
Want to try it yourself?
SAMSON provides downloadable computed trajectories in different formats, including:
The SAMSON file includes both the open and closed conformations, intermediate conformations, and metadata enabling replay and inspection of the full transition. A visual snapshot shows the full trajectory within the SAMSON Document View:

Please note: These trajectories haven’t been experimentally verified, but they provide a solid starting point for further refinement and analysis. They illustrate molecular motion that would otherwise be difficult to model directly.
To learn more, read the full documentation page here: Computing the Opening Motion of the SARS-CoV-2 Spike.
Note: SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





