Modeling conformational changes in large biomolecular assemblies is a recurring challenge in structural biology and drug design. One protein that drew intense focus is the SARS-CoV-2 spike, which transitions between closed and open states to interact with the ACE2 receptor. While the structures of both states are experimentally resolved, navigating the intermediate states can be quite difficult.
To address this, the SAMSON platform provides a computed transition trajectory of the SARS-CoV-2 spike going from its closed form (PDB ID: 6VXX) to its open form (PDB ID: 6VYB). This can be incredibly useful for modelers interested in docking, antibody design, or visual studies of viral entry mechanisms.
The trajectory was generated using two SAMSON modules: the ARAP Interpolation Path module and the P-NEB (Parallel Nudged Elastic Band) module. These tools allow users to interpolate between two states and refine the resulting pathway, even when the initial and final structures have a different number of residues—a common problem in structural modeling.
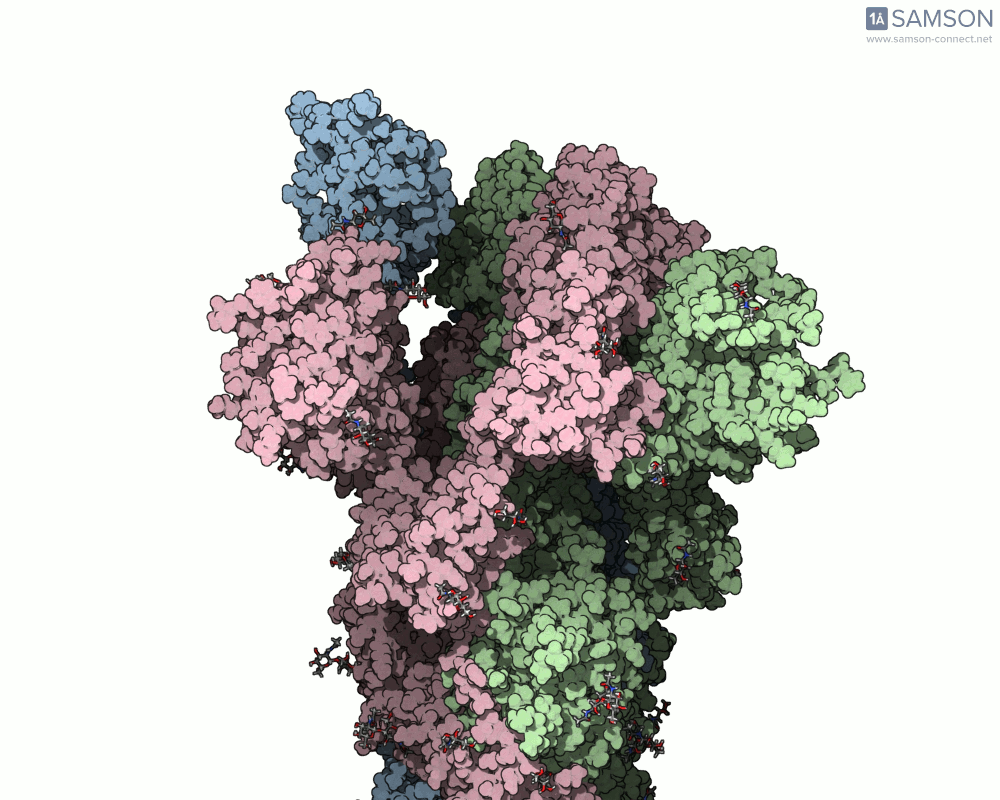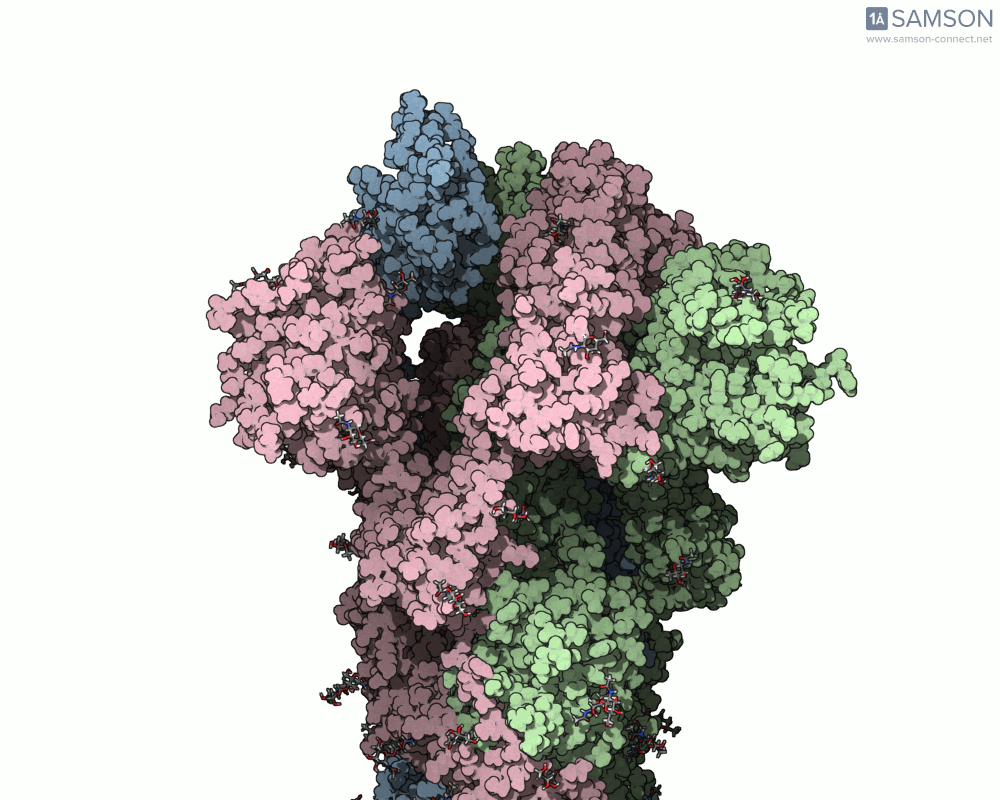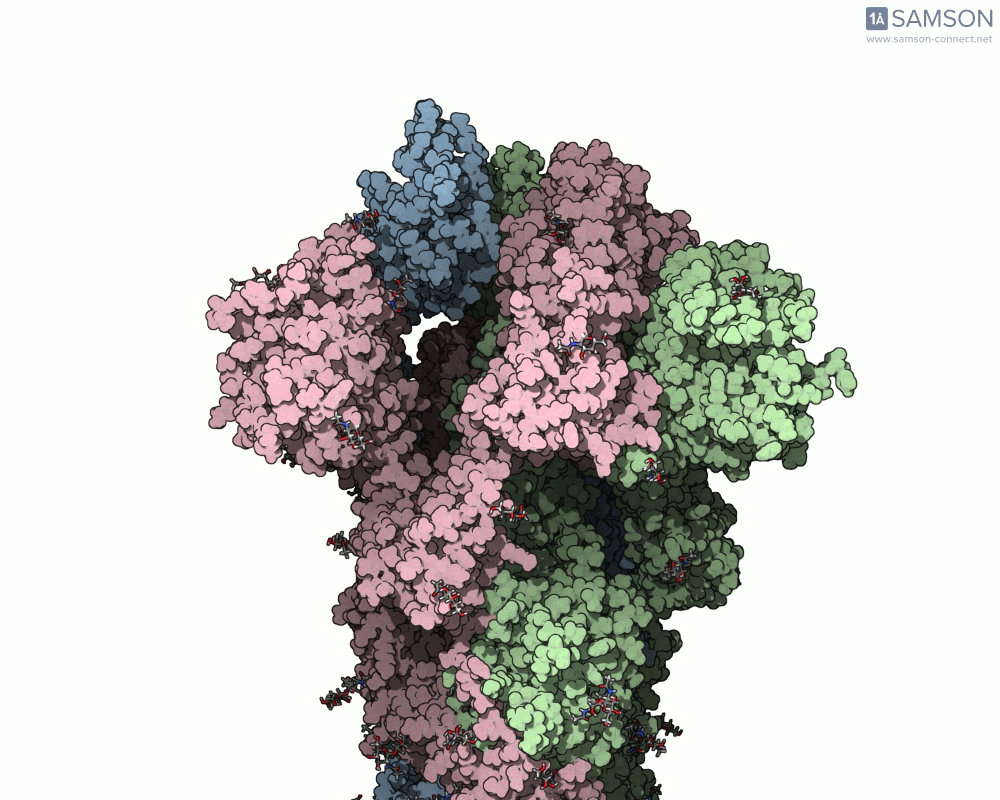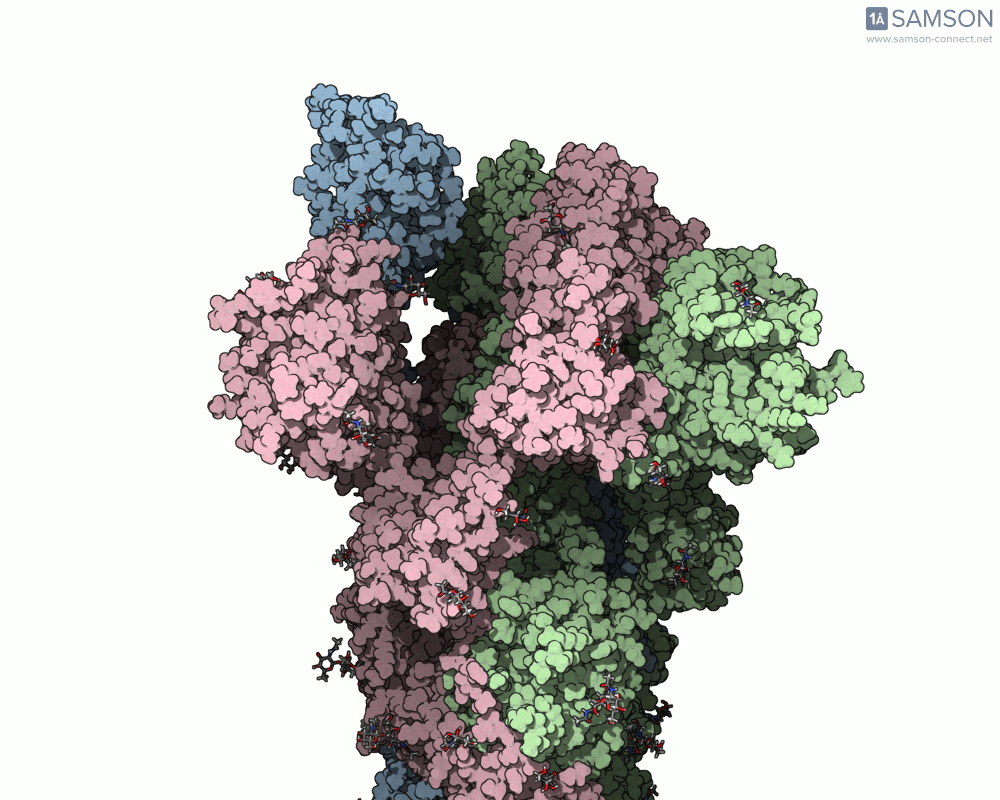
The motion reveals the receptor-binding domain (RBD) of one spike monomer lifting away from the trimer axis. This dynamic process is crucial for initiating viral attachment to host cells and is the principal target of neutralizing antibodies. Exploring this path provides insights that simple static structures cannot offer.
What you get
- GIF animations showing the spike from the side, angled, and top-down views.
- A downloadable PDB trajectory of intermediate conformations between the closed and open states (link to PDB trajectory files).
- A ready-to-use SAMSON document with all essential files integrated, including the computed path and conformations (SAMSON format download).

Why this matters
Animating this conformational transition can save you time when preparing simulations, developing structure-based hypotheses, or illustrating molecular mechanisms. Generating such transitions manually requires extensive setup, especially when the structures differ in residue count or require hydrogenation and minimization steps.
Using SAMSON’s automated pipeline, the process was executed with minimal manual intervention and completed in under 20 minutes on a laptop. The resulting animation is useful not only for scientific presentations but also for setting up further simulations or exploring how mutations could affect this motion.
To learn more about the methodology and the files provided, visit the full tutorial at this documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. To get started, download SAMSON at https://www.samson-connect.net.





