Structure prediction from NMR data can be very powerful, especially when working with protein-ligand complexes in solution. But many molecular modelers hit a common wall: incomplete or ambiguous methyl assignments. When multiple methyls are observed in a NOESY spectrum but can’t be assigned with certainty, the number of possible structure models grows quickly.
Fortunately, the NMR2 extension in SAMSON offers a way to deal with this by allowing partial methyl assignments. This can help guide the structure calculation process without over-restricting it, reducing meaningless permutations—and saving you hours of computational time and manual effort.
Why Are Partial Methyl Assignments Helpful?
In typical NMR workflows, you may only know that two methyl groups are on the same residue, or that a methyl must belong to a certain type of amino acid (e.g., ILE or MET), but not the exact position. Instead of discarding this limited but useful information, NMR2 allows you to incorporate it as a partial assignment.
This information can be crucial to make the search space more manageable when predicting the structure of protein-ligand complexes. It doesn’t eliminate possibilities entirely—just narrows them down.
How to Enter Partial Methyl Assignments in NMR2
The NMR2 interface in SAMSON includes a Partial assignment box. You can use it to define constraints such as:
|
1 2 3 |
same_res = M1 M2 same_res = M3 M4 |
This tells the algorithm that certain methyls live on the same residue. You can also assign residue types like this:
|
1 2 |
M1 = MET ILE |
These inputs guide the algorithm to test only valid permutations of assignments during structure prediction, improving performance and interpretability.
When Should You Use Them?
Here are some situations where partial assignments are particularly helpful:
- You observe peaks corresponding to methyls but can’t attribute all of them unambiguously.
- You’re dealing with a complex binding site with several possible contact residues.
- You have secondary data from other NMR techniques or mutagenesis suggesting certain methyl group associations.
Even if you only know that two methyls must come from the same residue, adding that information helps reduce combinatorial complexity.
What Happens If You Don’t Provide Them?
NMR2 defaults to testing all possible methyl combinations, and that number can become large very quickly. Without partial assignments, each methyl may be matched with any candidate, increasing the risk of ambiguous or low-quality structure predictions.
If speed, clarity, or computing cost is a concern, it’s worth revisiting what partial information you do have.
Visualizing the Outcome
After calculation, you can load the predicted structures into SAMSON and align them to inspect how the different predictions vary. Here’s an example after alignment and removal of pseudo-atoms:
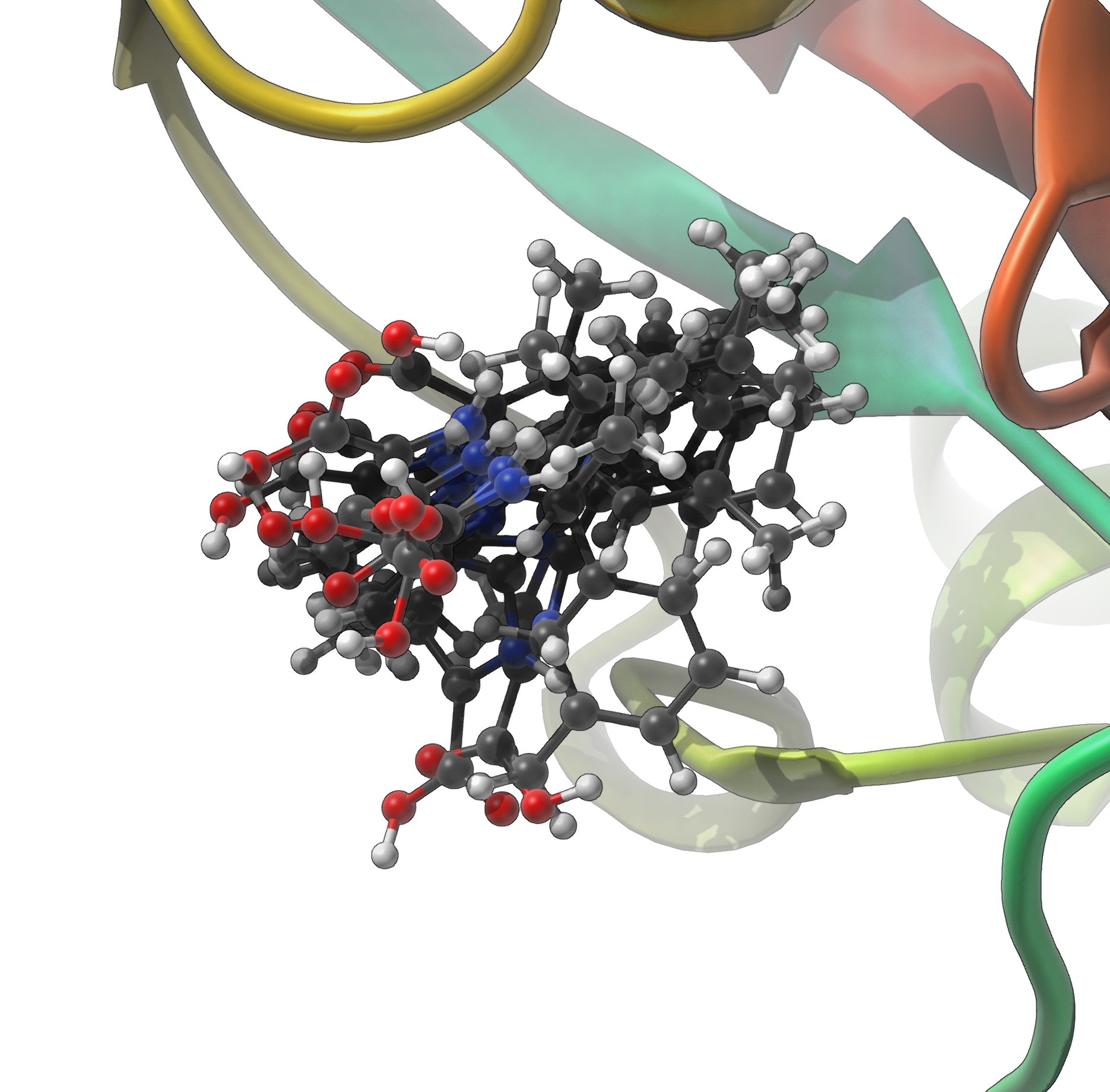
This step is especially informative when partial assignments reduce uncertainty: you’ll likely see a tighter cluster of predicted ligand poses.
For an interactive guide on how to use this feature, you can refer to the original documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





