Modeling molecular systems doesn’t always go smoothly. Sometimes, atoms need to connect. Other times, bonds have to break. In those moments, traditional molecular force fields—designed for static topologies—can get in the way. You might find yourself re-typing your molecules, manually reconstructing connectivity, or restarting your simulation just because a small structural change broke the whole system.
But what if your force field could handle all that automatically? That’s exactly what the Interactive Modeling Universal Force Field (IM-UFF) in SAMSON is for.
What Is Topological Flexibility, and Why Does It Matter?
In typical force fields, connectivity—the list of bonds between atoms—is fixed. Changing it means stopping the simulation and running bond detection algorithms again. This breaks the flow of interactive modeling, especially when building or editing molecular structures on the fly.
IM-UFF offers a more flexible alternative. It dynamically adapts molecular topology in response to user interaction. Grab an atom with your mouse and move it slightly: bonds stretch, but hold. Move it further: bonds may break, and new ones form based on proximity and energy minimization.
How It Works in Practice
Once you’ve selected IM-UFF in SAMSON and started the simulation, several things happen under the hood:
- The system constantly evaluates distances between atoms and adjusts bond orders, atom types, and connectivity in real time.
- Bond energy calculations automatically shift to reflect this dynamic state. IM-UFF uses a zero-energy reference point when atoms are non-interacting, making comparisons meaningful across different topologies.
- There’s no need to manually define bond types or atom types; the model takes care of that by interpreting continuous changes based on atomic positions.
This means that—as a modeler—you’re free to push, pull, build, delete, or connect atoms interactively, without worrying about breaking the model.
An Example in Motion
The short animation below demonstrates IM-UFF in action. Atoms are manipulated interactively, bonds stretch and break, and a new topology emerges naturally:
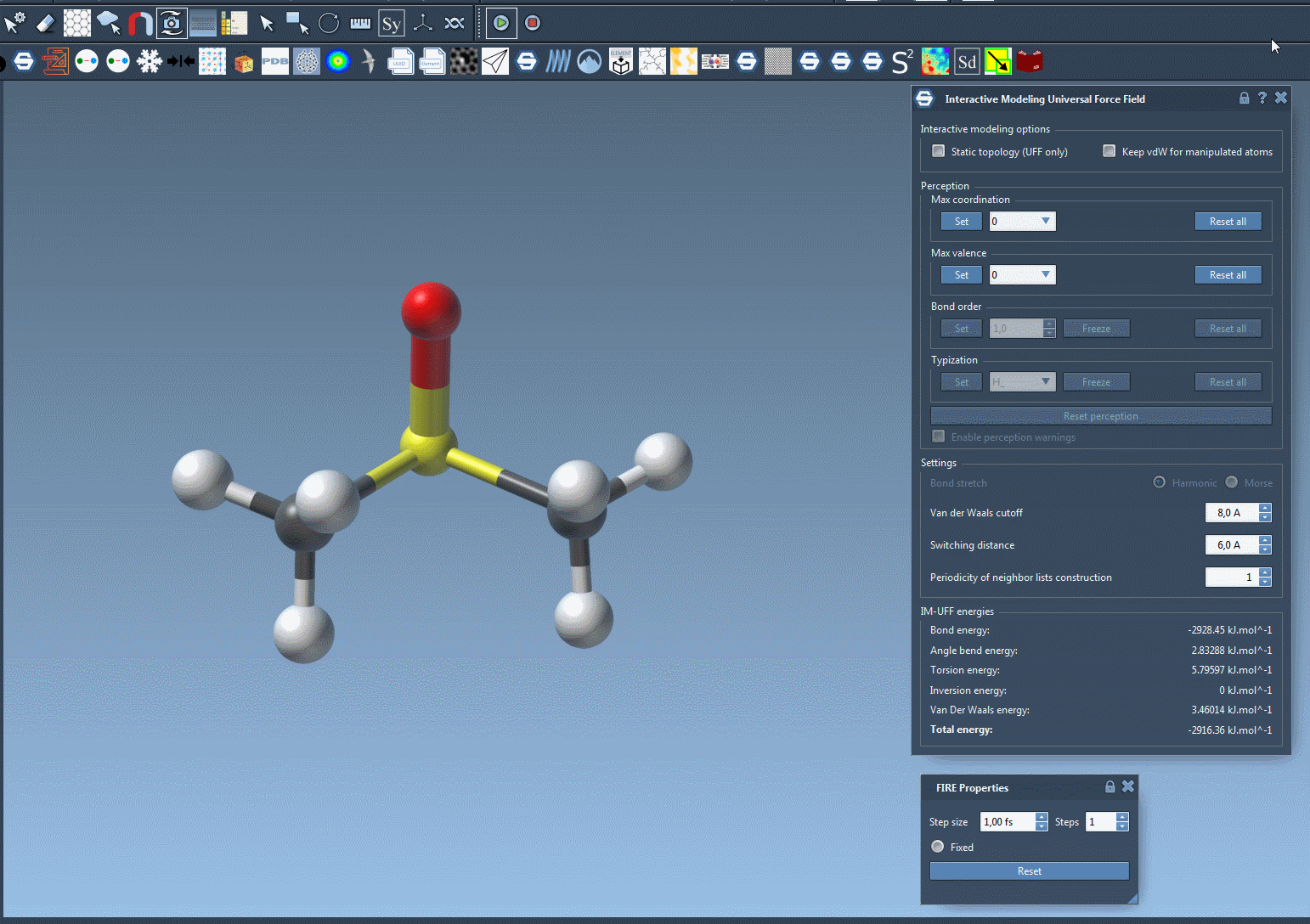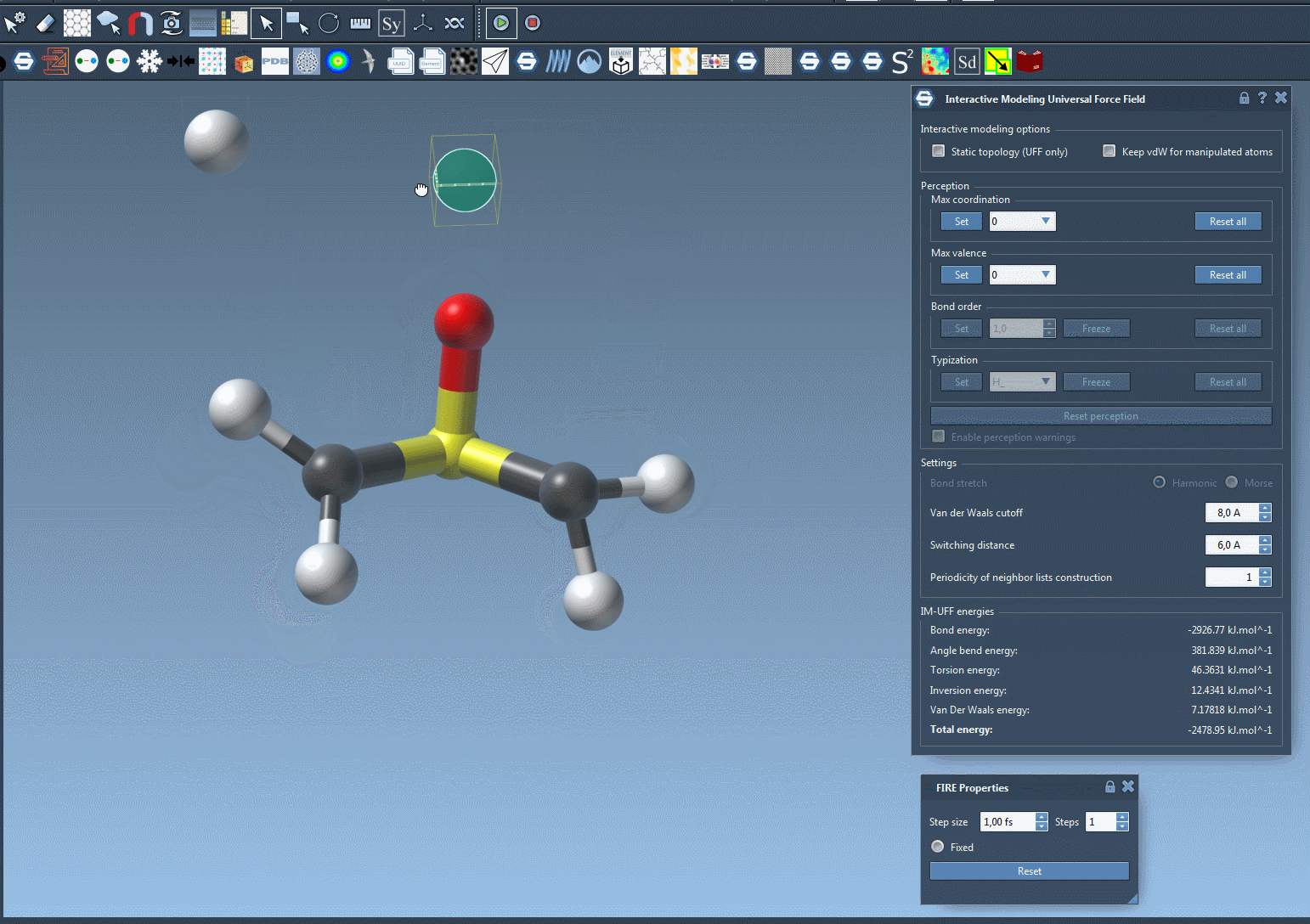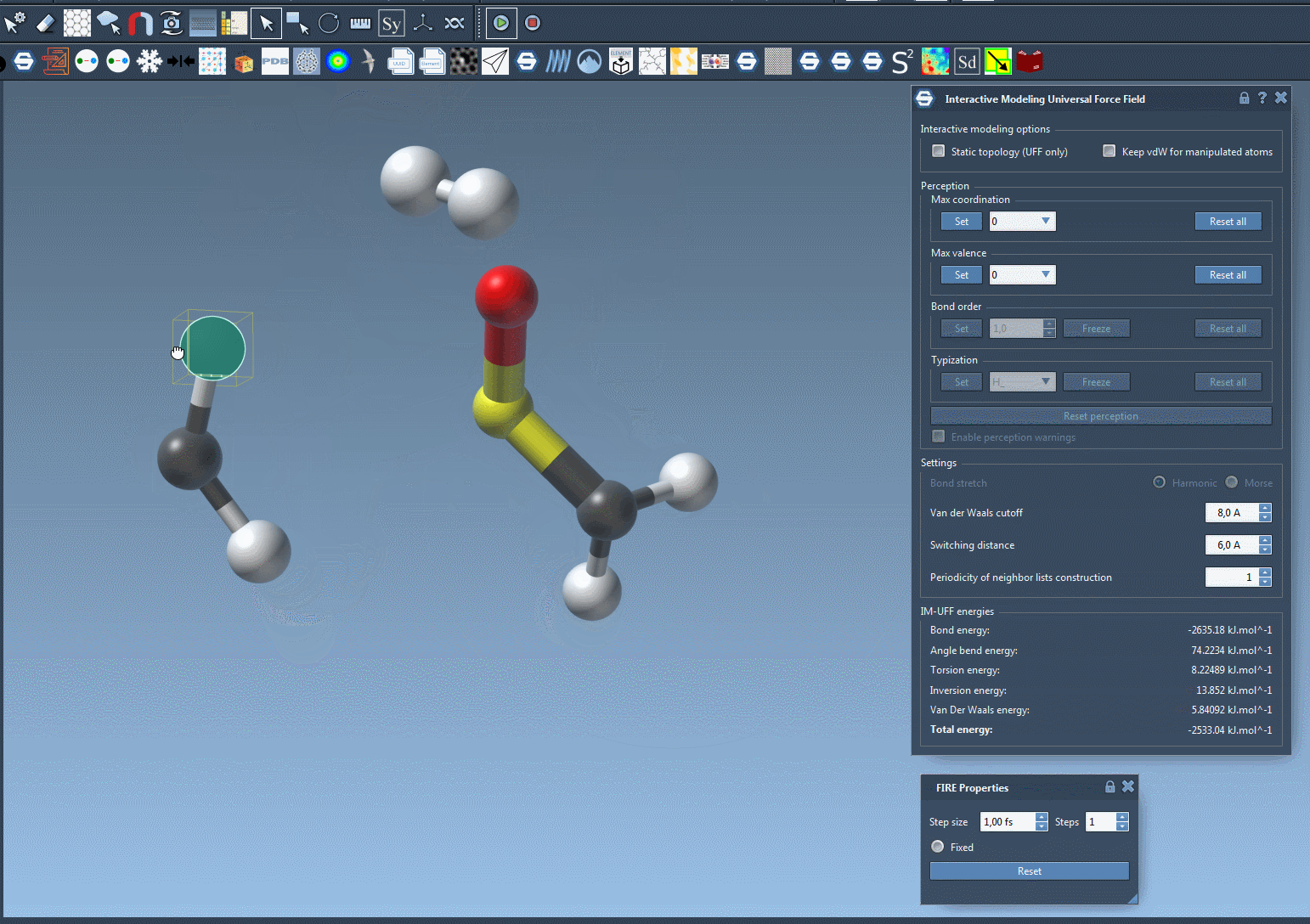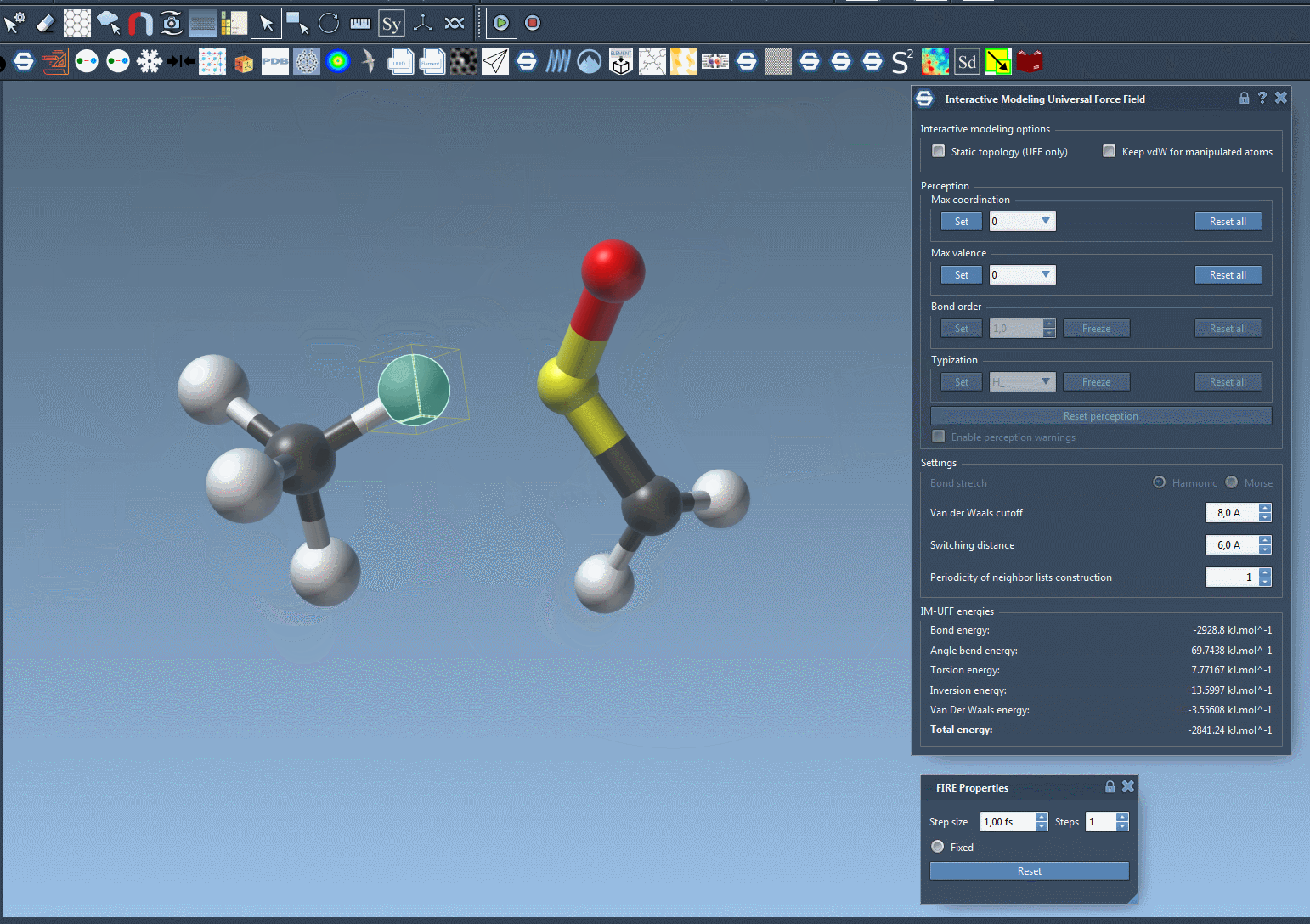
Notice how the system adapts smoothly, guided by inter-atomic forces. This is particularly useful when exploring pathways of molecular transformations or assembling molecules from fragments.
Tips for Better Control
More control is available through two key interactive options in the IM-UFF parameter window:
- Static topology (UFF only): Check this to turn off topology changes (same as standard UFF).
- Keep vdW for manipulated: Allow or disable van der Waals interactions for the atom you’re actively moving. Disable this temporarily when you’re trying to bring atoms very close together and vdW repulsion makes it difficult.
Use Cases and Benefits
IM-UFF is especially helpful in tasks like:
- Building novel molecules or materials from scratch.
- Manually correcting experimental structures by adjusting atom positions.
- Teaching concepts related to bond dynamics and molecular interactions.
Since the model is always adjusting bond order and topology dynamically, it frees you from the need to re-run setup steps. This can save significant time and reduce friction when iterating rapidly on structure design.
To learn more about how IM-UFF works and how to customize its behavior further—including automatic typization and energy term details—check out the full documentation at https://documentation.samson-connect.net/tutorials/uff/im-uff/.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





