One common challenge in molecular modeling is smoothly editing molecular systems while maintaining physical plausibility. Modifying a structure often requires deleting and recreating bonds manually, adjusting atom types, and reconfiguring the force field setup to reflect changes. These steps can become tedious—and risky—when you’re trying to preserve accuracy.
The Interactive Modeling Universal Force Field (IM-UFF), as implemented in the SAMSON Platform, offers a pathway to simplify these operations. Thanks to its ability to handle topological changes dynamically, IM-UFF allows you to simulate and interactively build or edit molecular systems, letting the force field make smooth adjustments as you go.
What does topology change mean in practice?
It means that when atoms move far enough apart, IM-UFF can break their bonds. Similarly, if atoms come close enough together, new bonds may form—automatically. During these transformations, atom types and bond orders are updated continuously, reflecting the new molecular context. No need to pause a simulation, update parameters, then restart.
To demonstrate this, try the following steps in SAMSON:
- Open or build a molecular system you’d like to edit or simulate.
- Go to Edit > Simulate > Add simulator (or press
Ctrl+Shift+M/Cmd+Shift+M). - Select Interactive Modeling Universal Force Field from the list of interaction models.
- Uncheck the Static topology (UFF only) option to enable dynamic bonding behavior.
- (Optional) Uncheck Keep vdW for manipulated to reduce repulsion when dragging atoms.
- Click Edit > Simulate > Start to begin the real-time modeling session.
Try this with your model
Move an atom slowly—its bonds will stretch, but stay intact. Now drag it further. At some point, the bond will break naturally as the energy becomes too high to sustain the connection. Bring the atom near another center, and if conditions are right, a new bond will form, updating the local molecular topology in a physically consistent way.
This behavior helps avoid one of the common sources of structural errors—manipulating molecules manually without underlying energy-based guidance. You can now prototype reactions, edits, or rearrangements without needing to stop and switch tools.
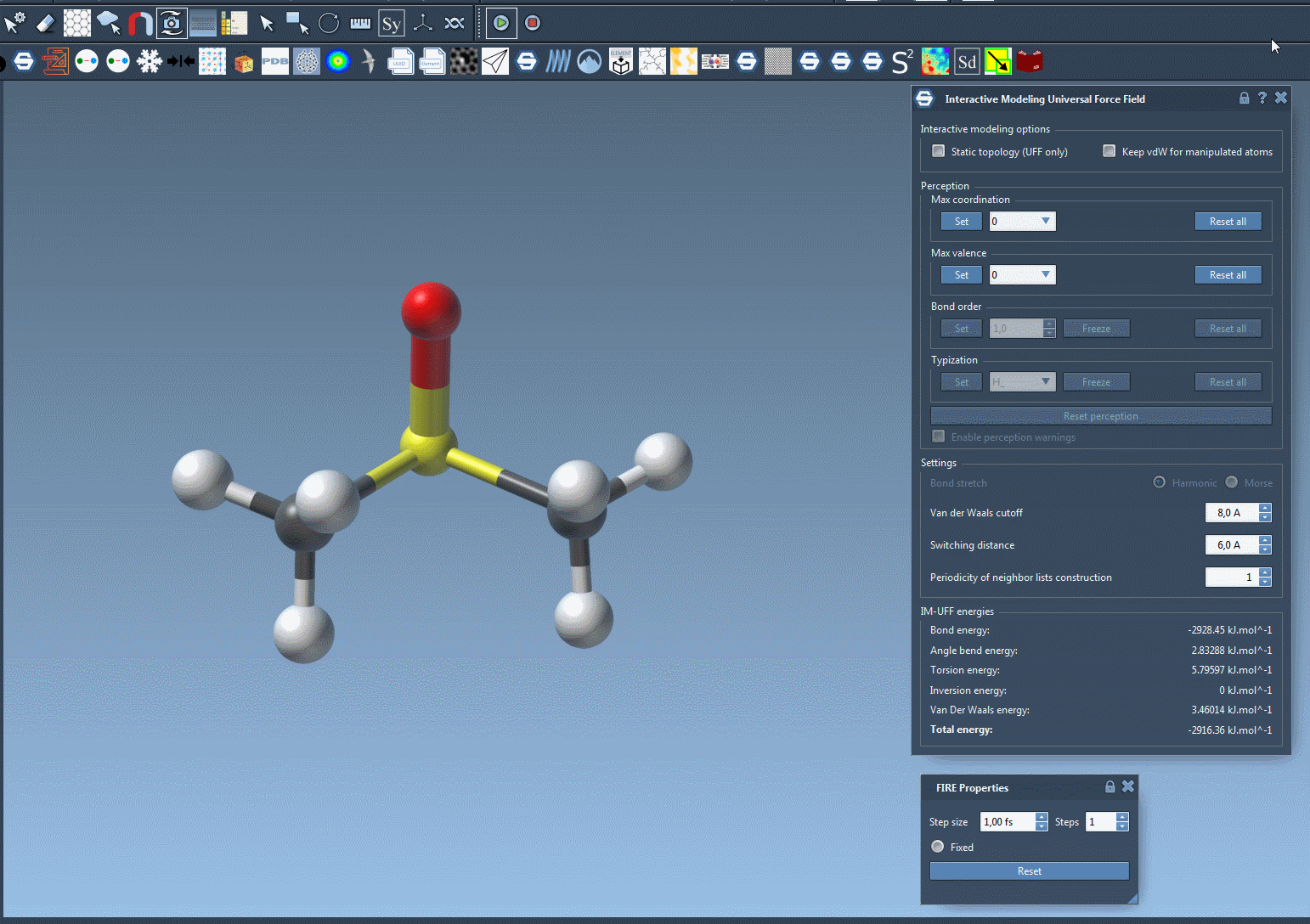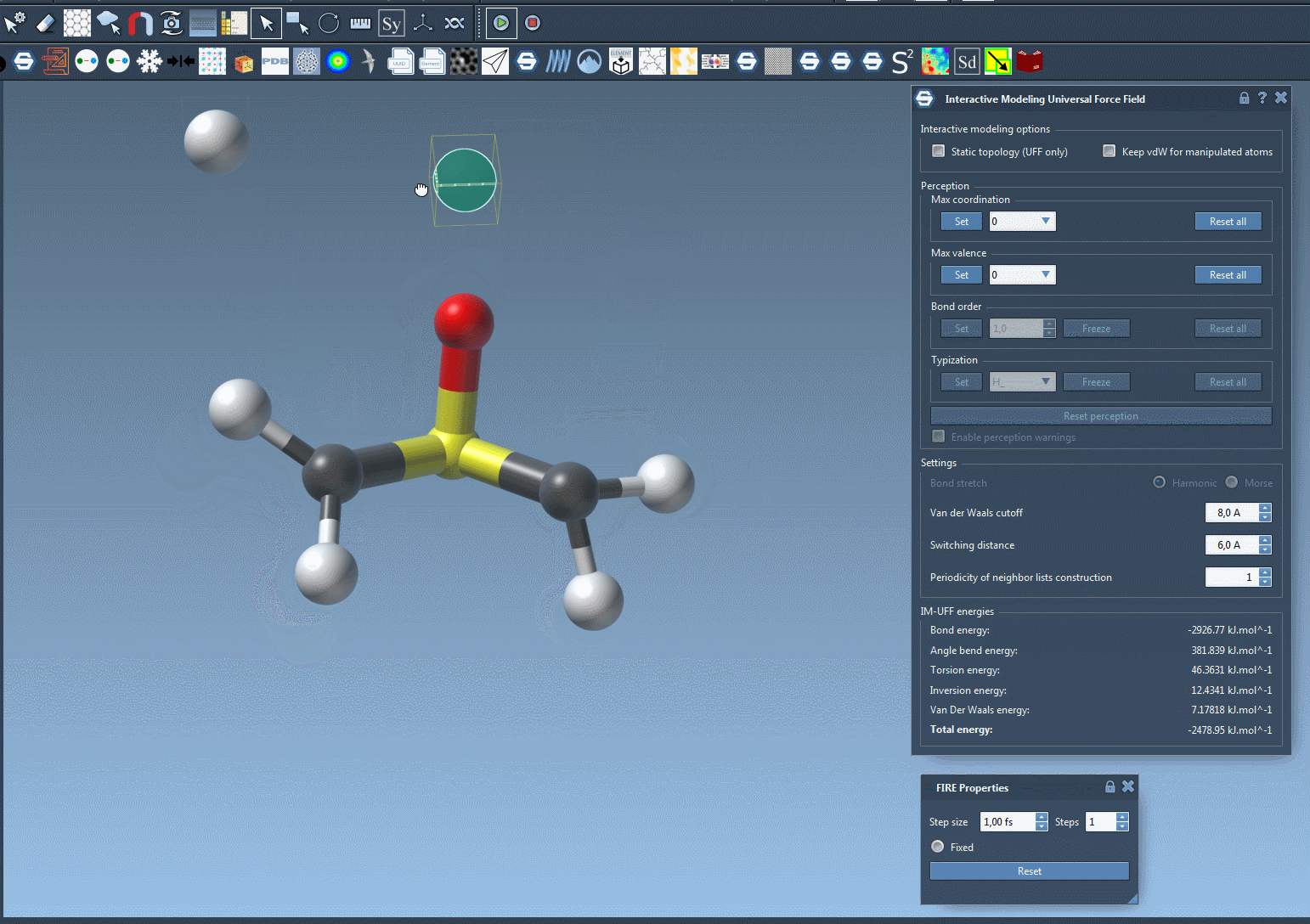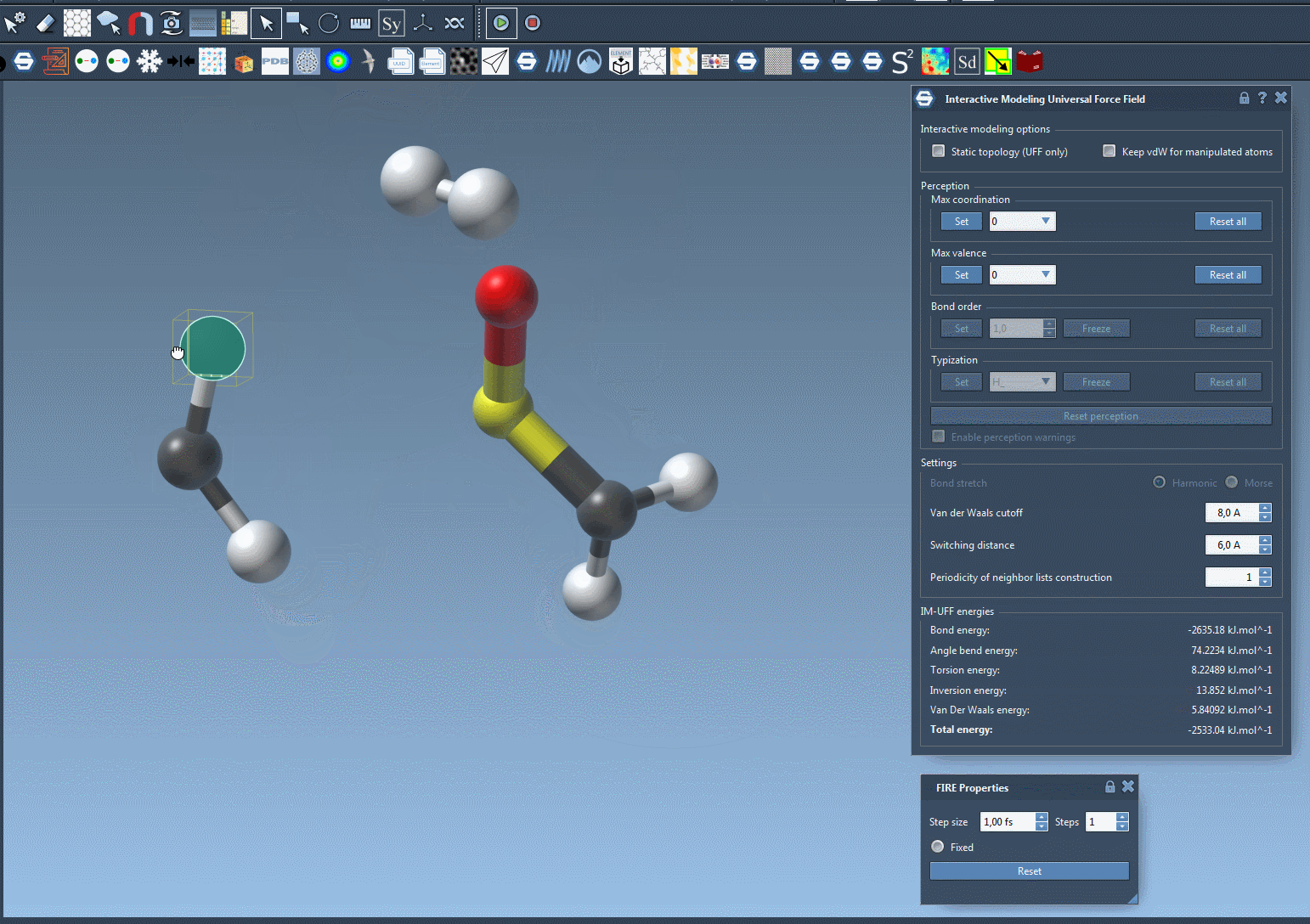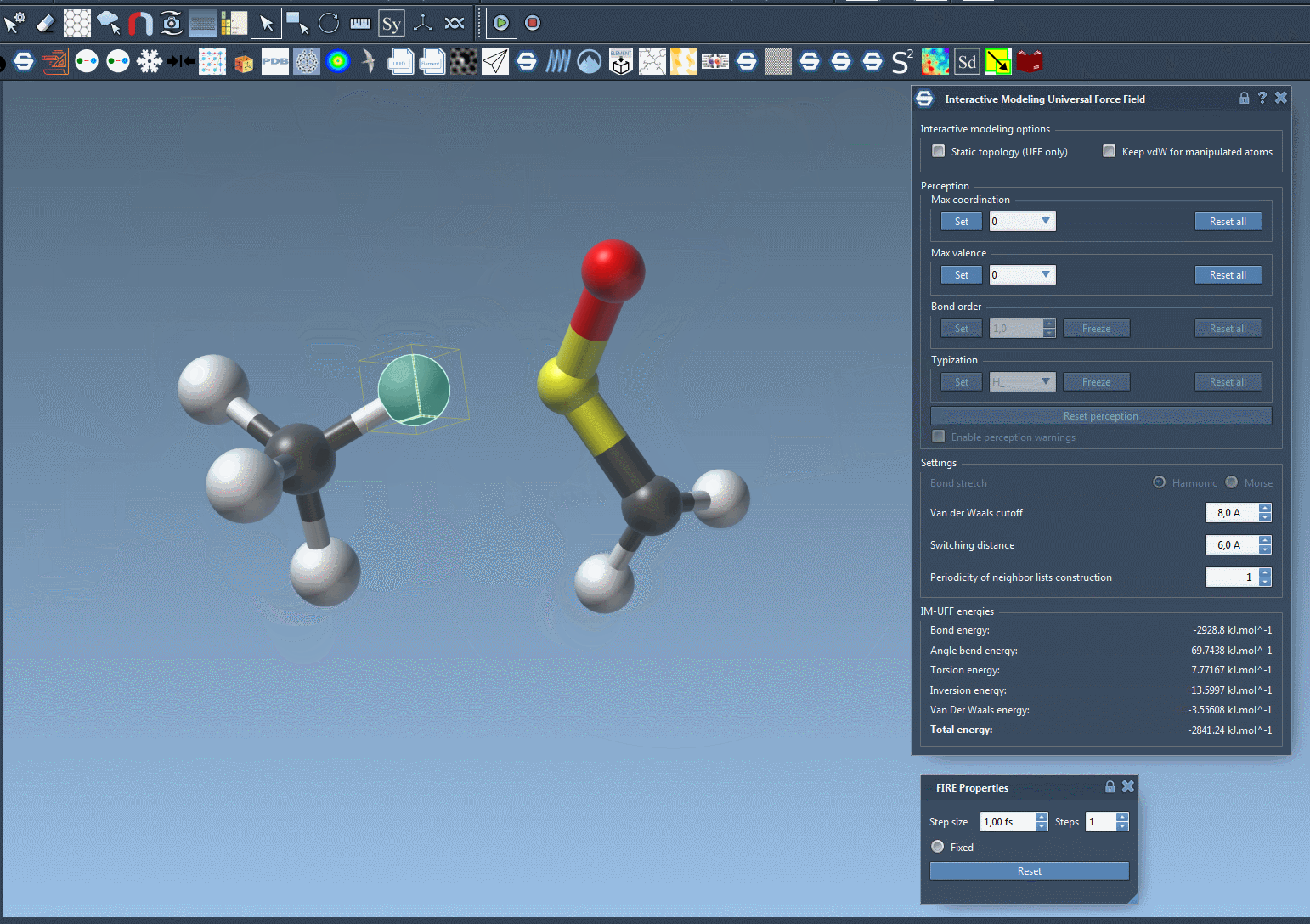
Why does this matter?
Whether you’re modeling hypothetical molecules or setting up educational demonstrations, being able to deform and reconfigure molecules on-the-fly makes the design loop much smoother. IM-UFF also avoids discontinuities in the energy landscape by using continuous bond orders and typization values, which results in more realistic visual and simulation responses as you edit.
You don’t need special configurations—IM-UFF works straight away as long as static topology is disabled. And for finer control, it allows you to set a few key parameters, such as maximum coordination and valence, that control how flexible the model is when changing bonding patterns.
Want to go further?
If you’re looking for a balance between control and automation in your interactive molecular modeling, this feature of IM-UFF is worth exploring. It helps keep your structures physically sound while giving you the freedom to explore different bonding scenarios in real time.
Learn more in the IM-UFF documentation.
Note: SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON here.





