When comparing protein structures for molecular modeling or drug design, global superposition is often just the first step. Sometimes, however, the real interest lies in a specific region—like a flexible loop, an active site, or a conserved domain. Aligning entire structures can mask these local differences, leading to misleading conclusions or mispositioned residues in downstream modeling.
This is where region-specific structural alignment becomes a valuable approach. In the SAMSON Protein Aligner, you can superimpose proteins based on selected residues only—a feature that offers more control over alignment for tasks requiring local precision.
Why Align by Region?
- Study subtle conformational changes in loops or binding sites.
- Focus on homologous regions conserved across diverse structures.
- Align flexible domains independently of the rest of the structure.
- Improve accuracy of region-specific modeling workflows (e.g., loop modeling, docking).
How It Works in SAMSON
Here’s how to perform a region-specific alignment using SAMSON’s Protein Aligner.
1. Load Your Proteins
As an example, consider aligning two hemoglobins: 1DLW and 1RTX. Open Home > Fetch, input their PDB codes, and load them:

2. Inspect the Global Alignment
With the Protein Aligner opened (via Home > Align), you can first attempt a global alignment. This gives you a general overview, but be aware that local regions may remain misaligned—like the pink N-terminal alpha-helices seen below:
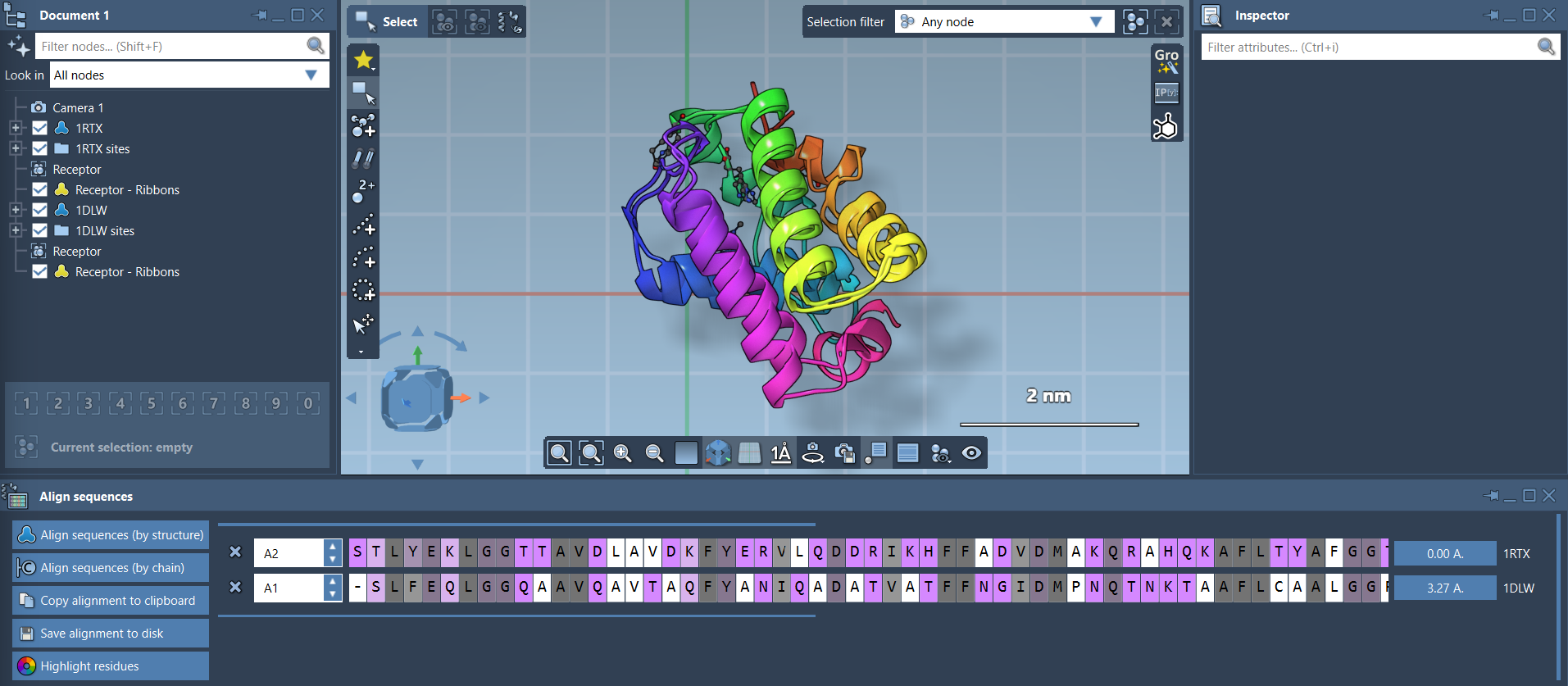
3. Select a Specific Region
Let’s say these N-terminal helices are your focus. Select the first 20 residues of each sequence directly from the Protein Aligner interface. You can use Shift for consecutive selections. Once selected, the alignment button next to the selection becomes enabled.

4. Align Only the Selected Residues
Click the alignment button (e.g., labeled 0.0 Å or showing an RMSD) next to the selection:

Only the N-terminal helices are now structurally superimposed, making it easier to compare their conformations independently from the rest of the protein.
Tips for Better Visualization
- Use Visualization > Visual model > Ribbons to clarify the structural features.
- Create separate ribbon models per protein to color them differently—for easier comparison.
- Use Highlight residues to visualize conserved properties such as polarity or similarity when available.
Final Thoughts
Region-specific alignment in SAMSON gives you granular control, ideal for cases where detailed structural analysis is needed. Whether you’re comparing flexible domains or aligning active sites only, this approach can provide clearer insights than global alignment alone.
Learn more in the full documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





