For many molecular modelers, the initial stages of a simulation can often be frustratingly slow. Before any meaningful analysis can begin, structures must be relaxed and optimized — and this step can become a bottleneck, especially for large or flexible systems. If you’ve relied on the steepest descent method for geometry optimization, you might have noticed that it’s not always the most efficient option, particularly when dealing with collective molecular motions.
This is where the FIRE Minimizer — the Fast Inertial Relaxation Engine — can make a difference. Included as a standard SAMSON Extension, FIRE offers a substantial speed-up in convergence, without making changes to your molecular model or requiring manual tweaking.
Understanding the Problem
During geometry optimization, structures are adjusted to reduce their potential energy. This process helps prevent artifacts that arise from unrealistic bond lengths or atomic clashes. The steepest descent algorithm, while intuitive, often requires small steps and a large number of iterations to handle motions that involve multiple atoms moving together (collective motions).
In contrast, FIRE combines aspects of molecular dynamics (momenta) with minimization logic, allowing for larger and more productive steps during optimization. The result — faster convergence with fewer steps — is particularly useful when preparing systems for simulation workflows or structural refinement.
FIRE vs Steepest Descent: A Visual Comparison
The difference in behavior becomes clear when watching both algorithms in action. The FIRE Minimizer produces smoother and faster relaxation over time, especially in complex starting configurations:

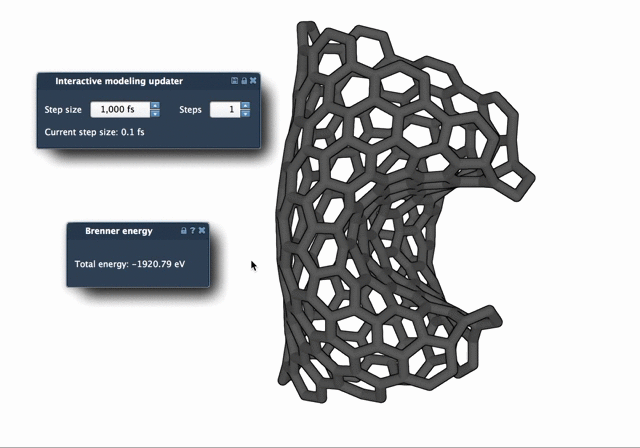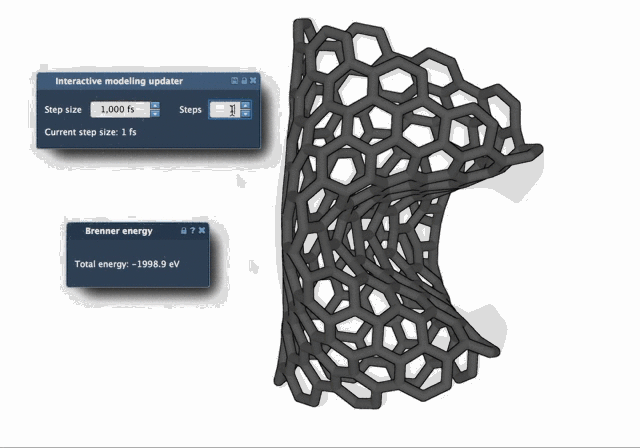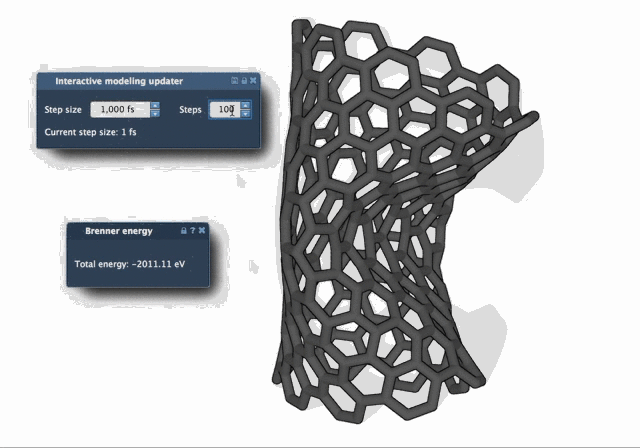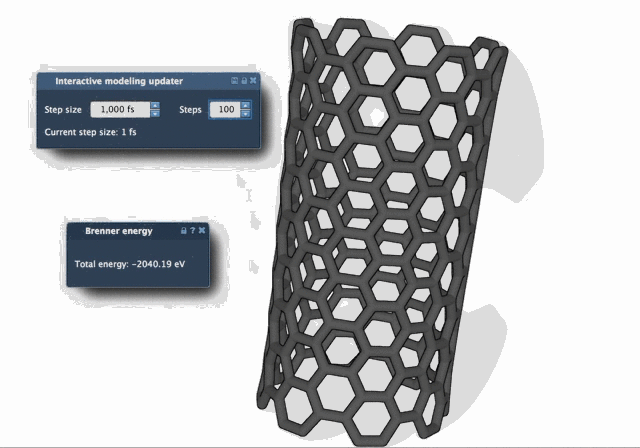
When to Use FIRE
- During initial clean-up of molecular structures from experimental sources.
- Before running molecular dynamics or quantum simulations.
- When the aim is faster convergence with fewer optimization steps.
- In workflows that involve iterative modeling and refinement (e.g. NMR structure fitting).
Getting Started with FIRE in SAMSON
To begin using the FIRE Minimizer in SAMSON:
- Log into SAMSON Connect.
- Install the FIRE Minimizer Extension.
- Restart SAMSON.
- Load your molecular model and add a simulator through
Edit > Add Simulator. - Choose your preferred interaction model and select
FIREin the State Updaters list.
Fine-Tuning FIRE Settings
The FIRE Minimizer is simple, but a few adjustable settings can provide additional control:
| Setting | Description |
|---|---|
| Step size | Initial integration step for minimization. |
| Steps | Number of FIRE steps before updating the viewport (visual feedback frequency). |
| Fixed | Optionally keeps the step size constant. |
For users who adjust atoms during minimization, the Reset button is available to reset FIRE’s internal history, ensuring proper behavior in the next minimization round.
If you’re working with large macromolecules, flexible systems, or need rapid clean-up before simulation, giving FIRE a try can streamline your molecular modeling workflow.
To learn more, visit the original documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





