Dragging atoms to explore structural rearrangements is a common task in molecular modeling, yet it often leads to frustration when the topology inconsistencies break your simulation or generate unphysical structures.
If you’ve ever attempted to manually tweak a molecular structure only to have your editing session end with broken bonds or frustrating force calculations, there’s a good chance you’re relying on a force field that doesn’t handle topology changes well.
This is where the Interactive Modeling Universal Force Field (IM-UFF) comes in. Available in the SAMSON platform, IM-UFF allows you to smoothly manipulate molecular structures while preserving their physics-based interactions, even as topologies evolve.
So, what’s the main issue?
Conventional force fields, like UFF, often assume a static topology where covalent bonds and atom types are fixed. In practice, this means that as soon as you start dragging atoms around, the system may push back aggressively, or worse, behave unrealistically if the structural integrity breaks down.
For researchers and modelers who want to interactively build, edit, and test molecular systems, this rigidity becomes a bottleneck. Why can’t we move atoms, break bonds, and form new ones as intuitively as we do on the whiteboard?
The solution: Interactive Modeling with IM-UFF
IM-UFF goes beyond UFF by enabling real-time topology updates. When combined with interactive manipulation (e.g., dragging atoms with your mouse), it handles changes such as:
- Bond formation and dissociation
- Change in bond orders
- Atom retypization in response to spatial configuration
This allows the molecular system to smoothly adapt as you explore different geometries or simulate structural responses to local modifications.
Here’s how to try it:
- Open a document with a molecular model in SAMSON.
- Add a simulator via Edit > Simulate > Add simulator (or press
Ctrl+Shift+M/Cmd+Shift+M). - Select Interactive Modeling Universal Force Field from the interaction models list.
- Uncheck both Static topology (UFF only) and Keep vdW for manipulated options (this setup enables full IM-UFF mode).
- Click Edit > Simulate > Start, and start experimenting by dragging atoms.
Try moving atoms short distances versus large displacements and observe how topological restructuring occurs dynamically. Bonds break and form based on atom proximity, driven by energy minimization and van der Waals interactions.
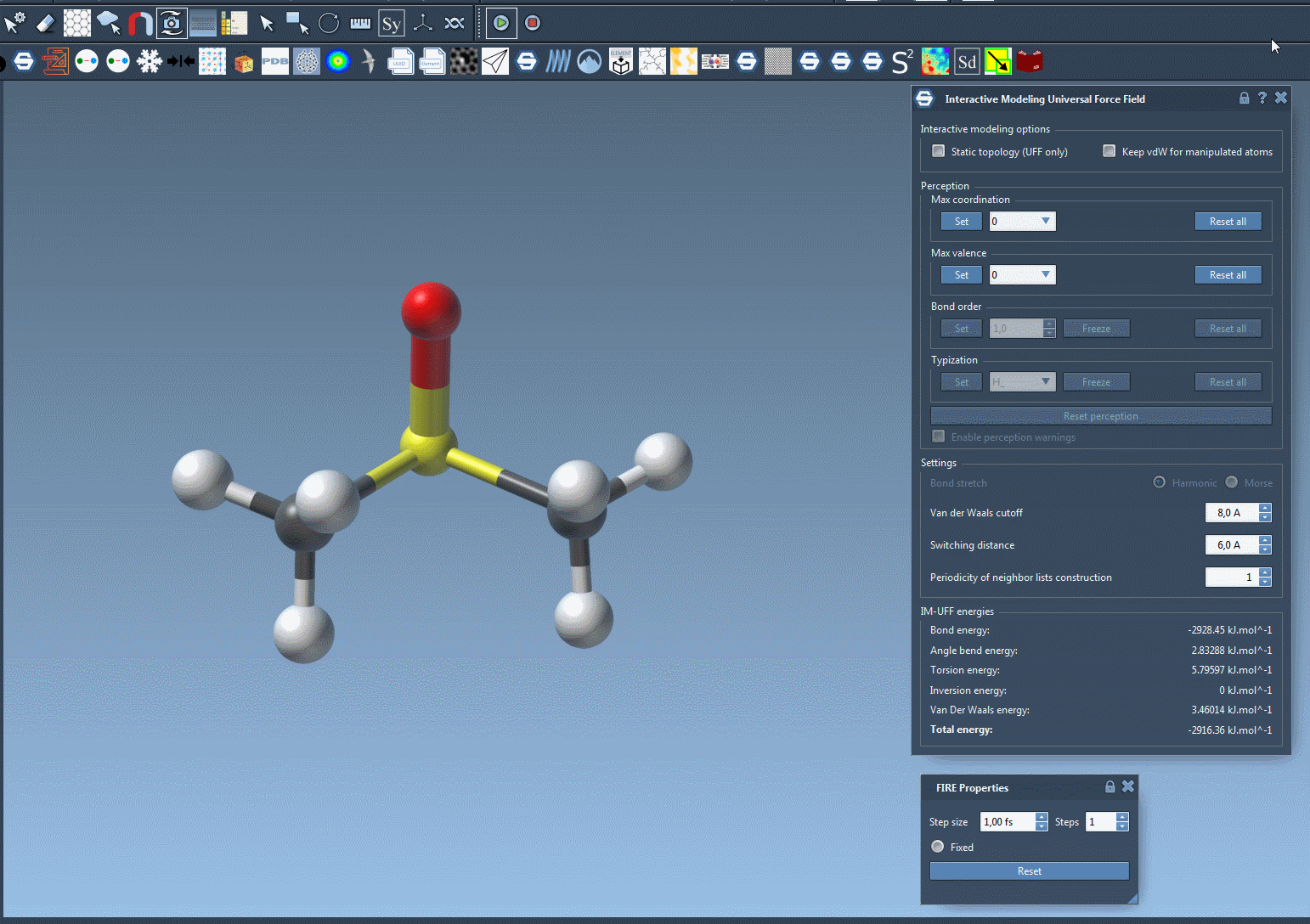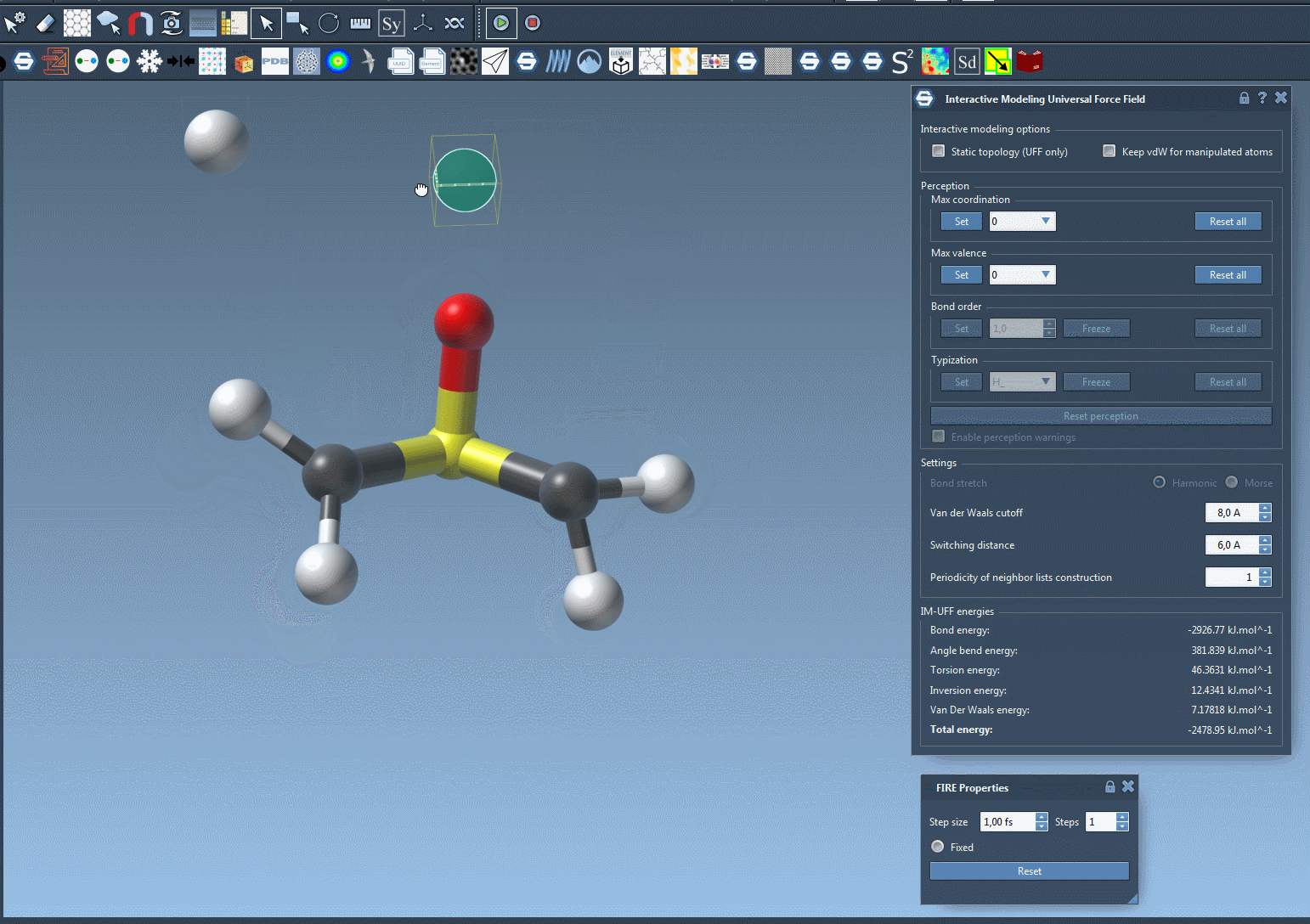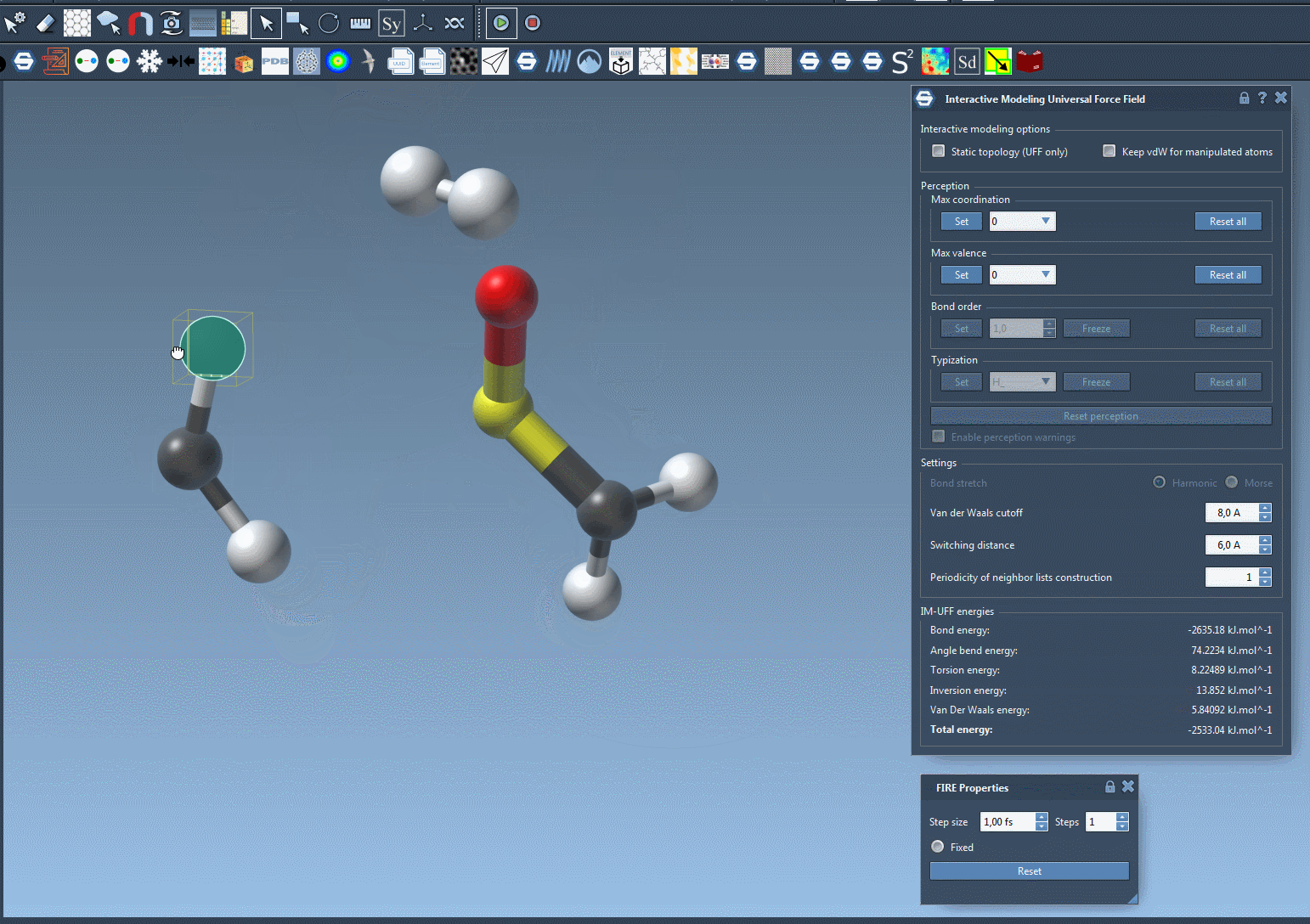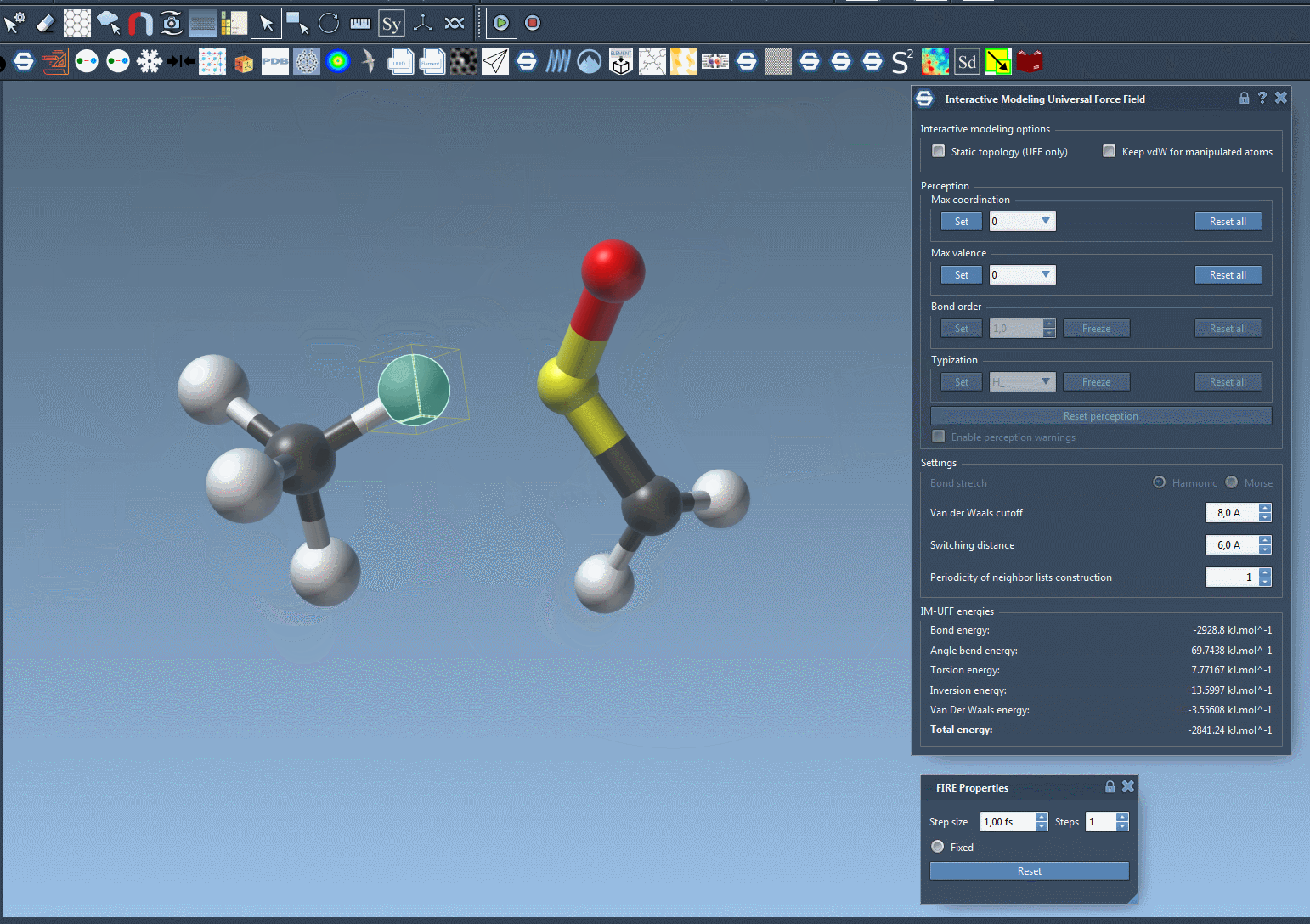
Making Manipulations Easier: Optional Controls
If you’re fine-tuning molecules and the van der Waals forces are preventing you from connecting atoms easily, just toggle off vdW consideration for manipulated atoms (via the Keep vdW for manipulated checkbox). This enables simpler docking or bond formation with minimal resistance during editing.
Summary
IM-UFF addresses one of the most persistent pains molecular modelers face: enabling intuitive, interactive adjustment of molecular structures while preserving realistic physical behavior. With real-time feedback and automatic typization, it streamlines the process of prototyping new molecular motifs, correcting structure issues, or simply exploring chemical possibilities.
To learn more about IM-UFF and step-by-step usage, visit the full documentation page here: https://documentation.samson-connect.net/tutorials/uff/im-uff/.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





