For many molecular modelers, preparing accurate 3D structures from lists of SMILES codes can become a repetitive and tiresome step. Whether you’re building a compound library, exploring analogs, or preparing systems for simulations, having a fast, reliable way to convert SMILES strings into 3D molecular structures is essential. Fortunately, SAMSON’s SMILES Manager, powered by RDKit, provides a streamlined workflow for this task.
In this post, we’ll look closely at one of the most sought-after functionalities of the SMILES Manager module: generating 3D structures directly from SMILES codes. If you’re dealing with hundreds — or even thousands — of molecules, this small but robust tool can save a significant amount of time and avoid errors from manually building structures.
What Is the 3D Structure Generator?
The main feature we’re covering here allows you to convert one or multiple SMILES strings (*Simplified Molecular Input Line Entry System*) into 3D molecular models. These 3D models can then be used immediately within SAMSON for property calculations, simulations, or exporting to other software.
How It Works
Start by importing your SMILES strings into the “Manage SMILES” tab in the SMILES Manager. You can do this by opening .smi or .txt files via the File menu (Open), or simply paste them into the table.
Once your molecules are listed, follow these steps:
- Select one or more molecules from the table.
- Click on the Export drop-down menu.
- Choose Selected SMILES string to Document.
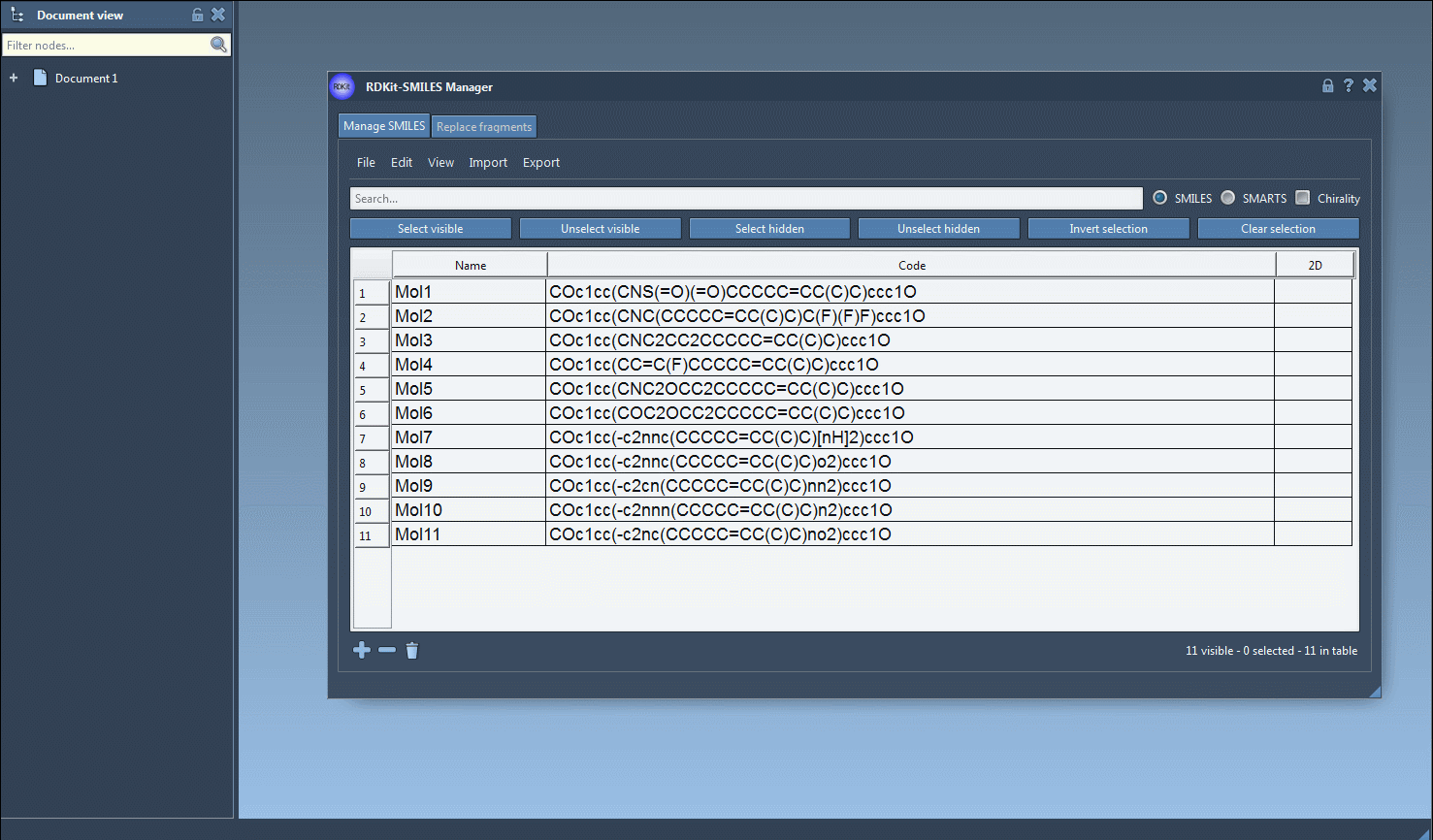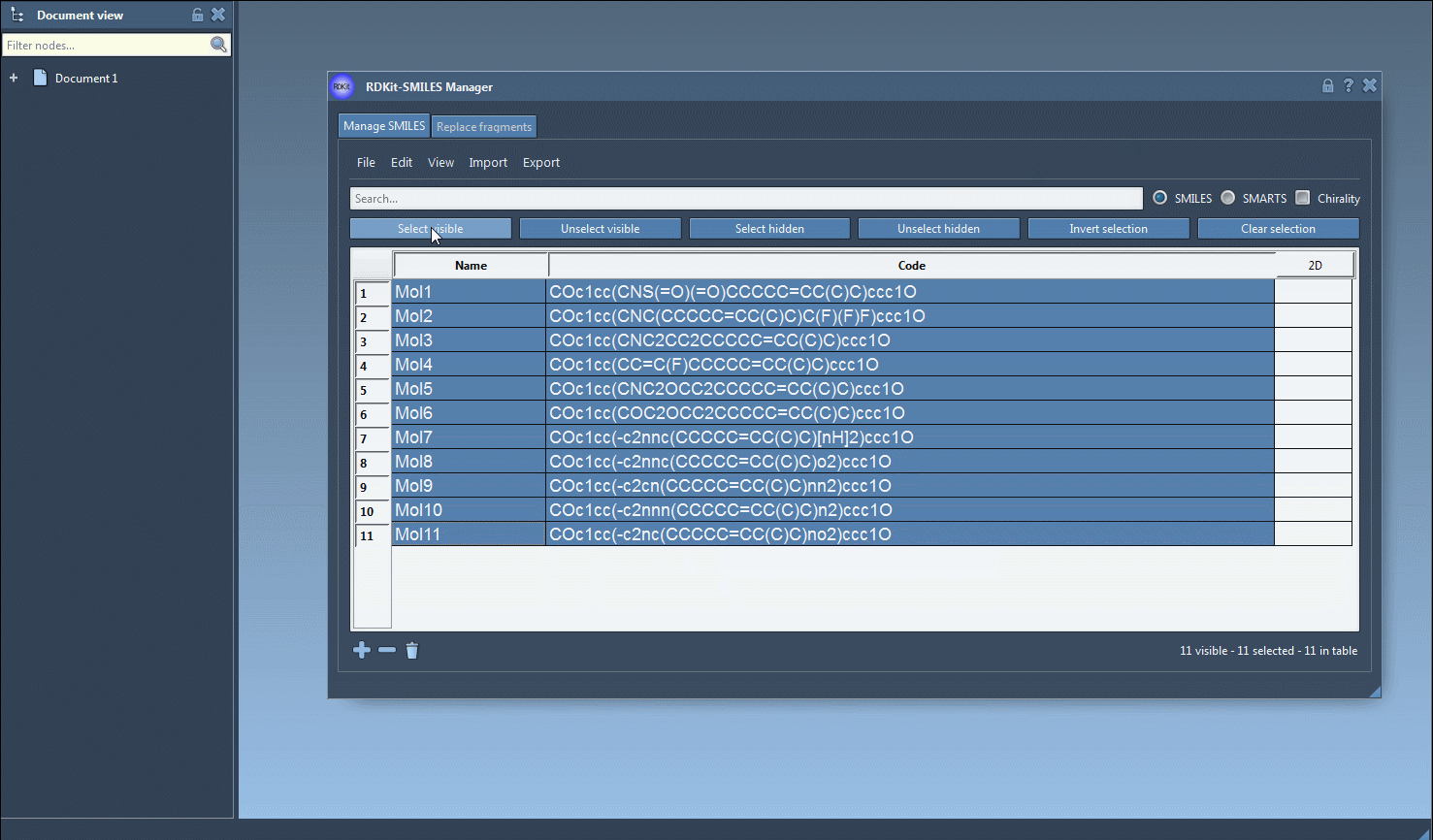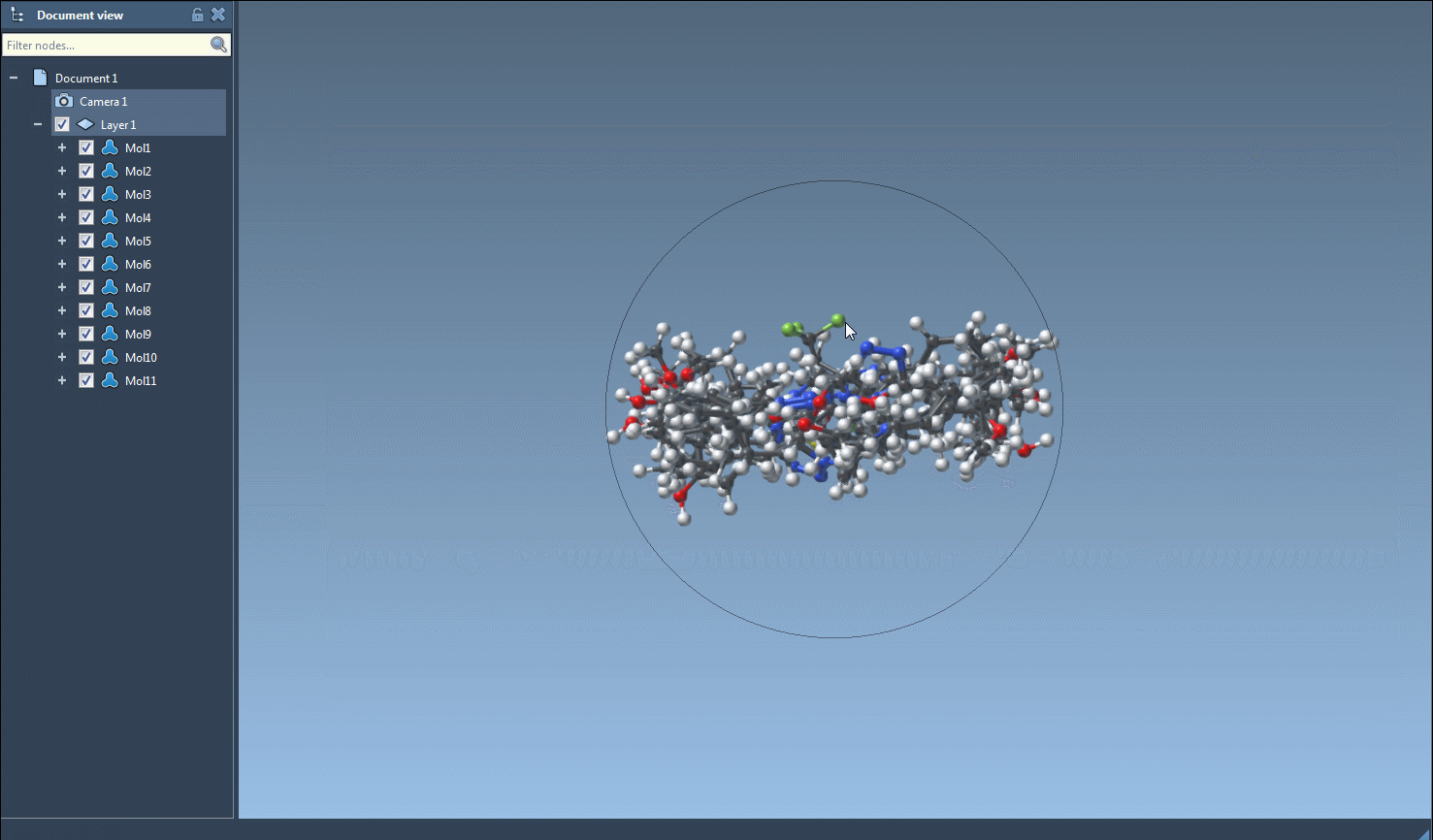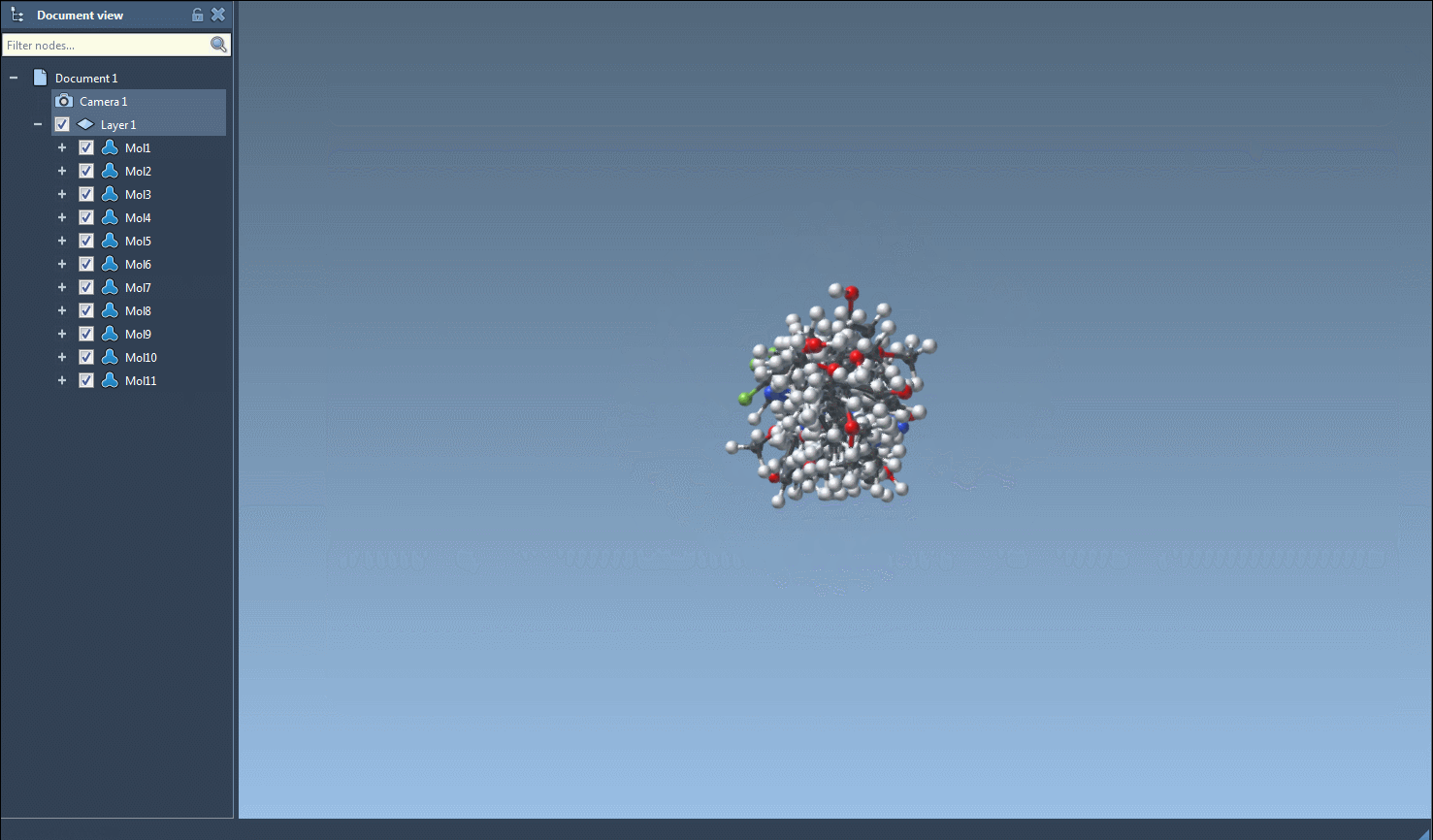
This will instantly generate 3D representations of the selected molecules and add them directly to your active SAMSON document.
You can also do this on a per-molecule level. Right-click any molecule in the table and select Generate 3D structure, or open the 2D depiction in a larger window and click on the Generate 3D structure button from there.
What Happens If a SMILES String is Invalid?
If the SMILES string is not correctly formatted or chemically invalid, the SMILES Manager will flag it. The corresponding line in the table will be highlighted, and instead of a 2D depiction, you will see an error image. This quick feedback helps navigate large SMILES datasets and spot problematic entries before proceeding.
Why It Matters
Manually converting lists of SMILES strings into 3D structures often requires juggling multiple tools, scripts, or web APIs — and occasional errors can lead to invalid molecules or incorrect geometries. With the SMILES Manager, the transformation is local, integrated, and immediate. This makes it easier to:
- Build 3D libraries for docking or simulation.
- Quickly screen diverse chemical scaffolds.
- Validate molecule integrity with visual inspection.
Bonus: Visualization and Export Options
Once 3D structures are generated, you can navigate among them using the SAMSON interface, zoom in/out, and even save visualizations as images for presentations or reports. Combined with 2D grid image exports from the same tool, this helps document your datasets more effectively.
Ready to Try It?
To learn more about this function and the other capabilities of the SMILES Manager, visit the full tutorial: SMILES Manager Documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON here and get started today.





