Molecular modeling often involves watching how a ligand binds to or unbinds from a protein. But extracting only relevant atomic coordinates for simulations like free energy calculations can be tedious — especially if your tool exports everything indiscriminately. What if you only care about the ligand?
The Export Along Paths extension in SAMSON offers a practical way to export just the ligand (or any subset of atoms) as it moves along a predefined path — streamlining studies of binding, unbinding, and conformational transitions.
A Common Pain: Too Much Data
Imagine you’ve used a ligand path-finding tool to model how a molecule departs from a protein’s active site. You want to perform free energy calculations or visualize only the ligand’s journey. If your program outputs the entire system — thousands of atoms over dozens or hundreds of frames — the resulting data can be too heavy for downstream tools, and harder to interpret.
Export Exactly What You Need
With SAMSON’s Export Along Paths app, you can export just a selected subset of atoms, like a ligand or even a small molecular group, and skip everything else. Here’s how that works:
1. Select Your Subset
Use the Document view to find and select the specific molecule you’re interested in — for example, the ligand TDG in the Lactose permease system:

2. Add as Export Model
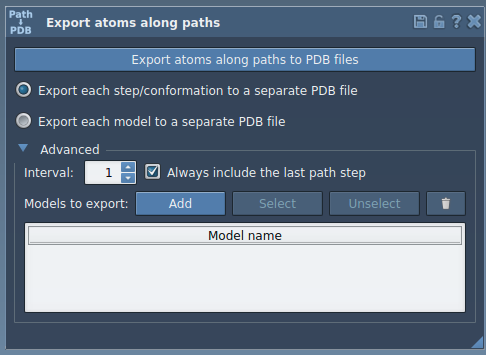
In the Export Along Paths app, open the Advanced panel and click Add. This defines a group of atoms to export. You can rename the model (e.g., “Ligand-TDG”) or define multiple subsets for more complex workflows.

You can also:
- Select/Unselect atoms from the subset dynamically
- Reset selection using the eraser icon
- Save several subsets (e.g., ligand + binding site)
3. Export Along the Path
Select the relevant path(s) in the Document view — these track how the ligand moves through space. Then initiate export:
- Choose export mode: single PDB with all frames, or one PDB per frame
- Set a file prefix and directory
- Adjust frame interval if needed
Why This Matters
Exporting only what you need not only reduces file size but also improves simulation efficiency and visualization focus. For workflows like free energy perturbation, you’ll often need structures along a reaction coordinate — not full systems. The Export Along Paths tool simplifies that.
Typical Use Cases
- Generating ligand-only PDBs along exit/entry paths
- Extracting backbone atom positions during a conformational change
- Building reaction coordinate files for umbrella sampling or metadynamics
- Creating meaningful visualizations without visual clutter
Careful data export may not be the flashiest part of molecular modeling, but it makes a real difference in productivity and clarity. If you haven’t used this export method yet, it may simplify your workflow more than you expect.
To explore more options and detailed instructions on path-based coordinate export, visit the original Export Along Paths documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON from https://www.samson-connect.net.





