Molecular modelers working with large biological systems like viral spike proteins often face the challenge of understanding conformational motions — especially when experimental data is limited to static states. This becomes particularly important for studying viral entry mechanisms, such as those involving the SARS-CoV-2 spike protein.
To help address this problem, the team behind SAMSON has shared an accessible workflow to simulate the large-scale motion of the SARS-CoV-2 spike protein from its closed state to its open, receptor-binding state. The motion isn’t purely theoretical — it’s based on known structures and computed using dedicated molecular design modules, enabling useful visualizations and further computational research.
Why simulate the spike transition?
Much of the SARS-CoV-2 spike’s function lies in its ability to switch from a closed to an open conformation. In its open form, it can engage the ACE2 receptor on human cells, initiating infection. Understanding and visualizing this transition helps researchers:
- Identify potential targets for antibodies or inhibitors
- Provide input samples for flexible docking or molecular dynamics simulations
- Explore structural landscapes even when no experimental transition trajectory is available
The trajectory in action
The SAMSON documentation includes a series of visual animations showing different views of the spike’s opening motion:
- Side view:
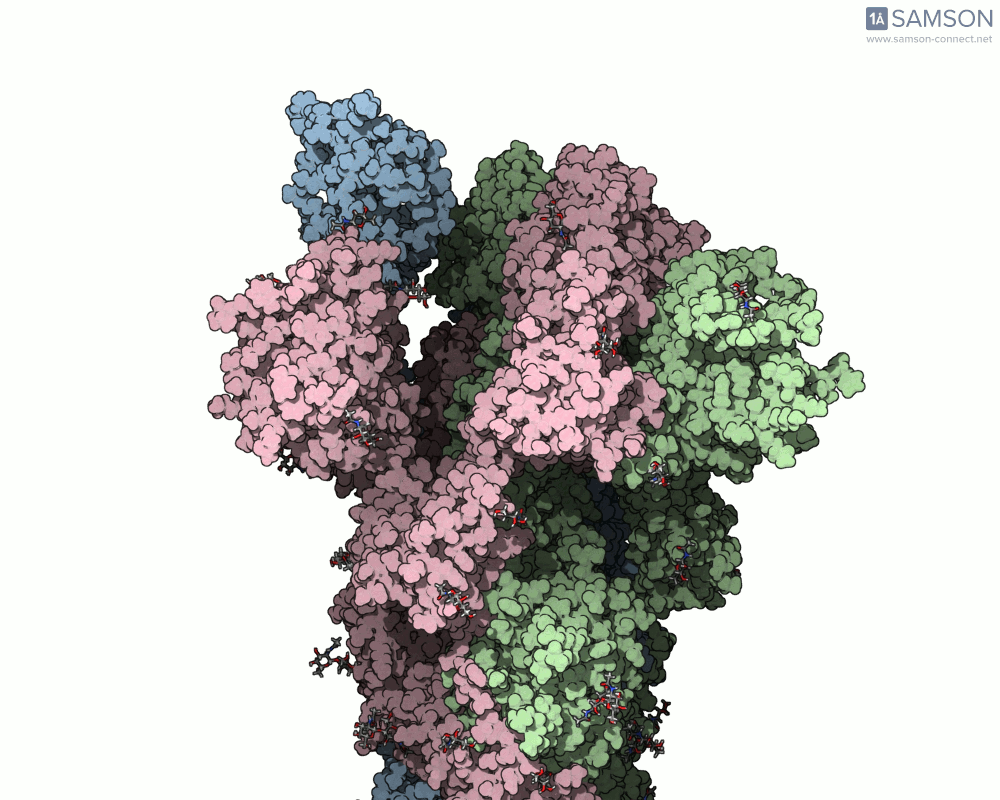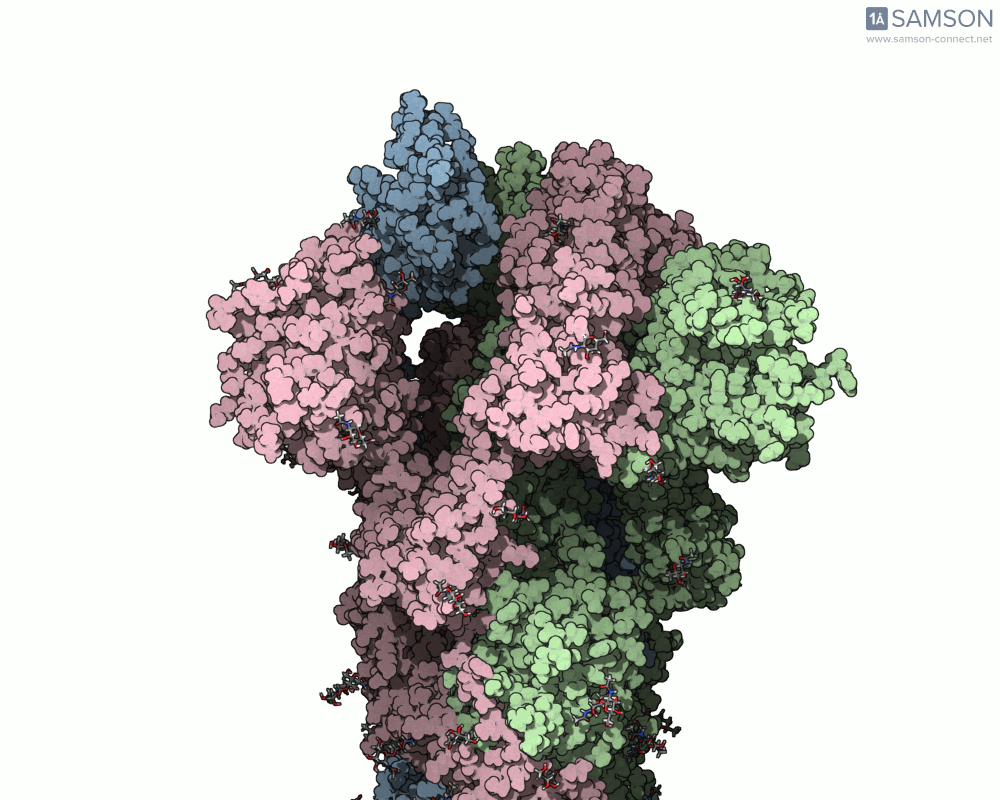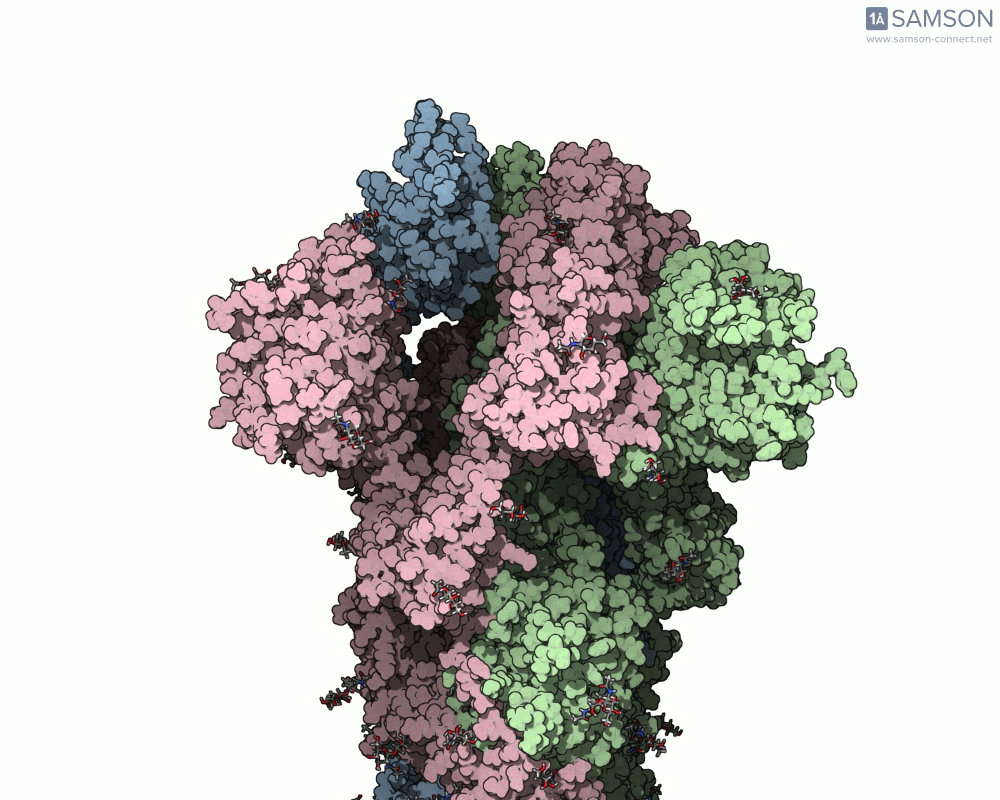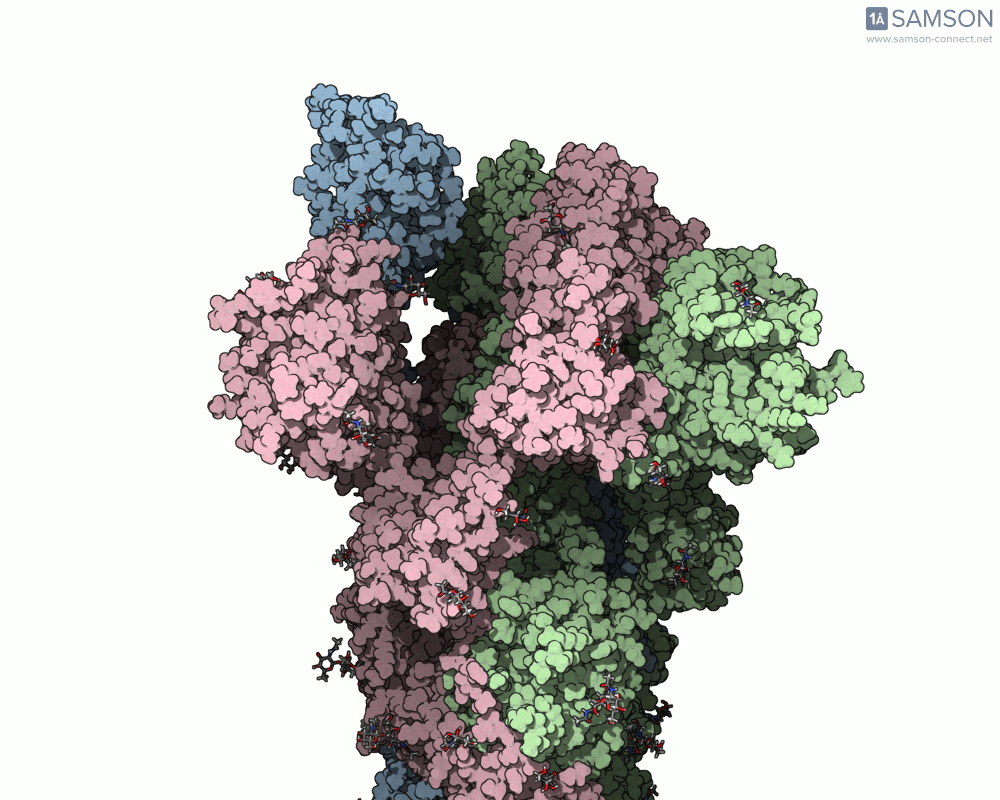
- Angled view:
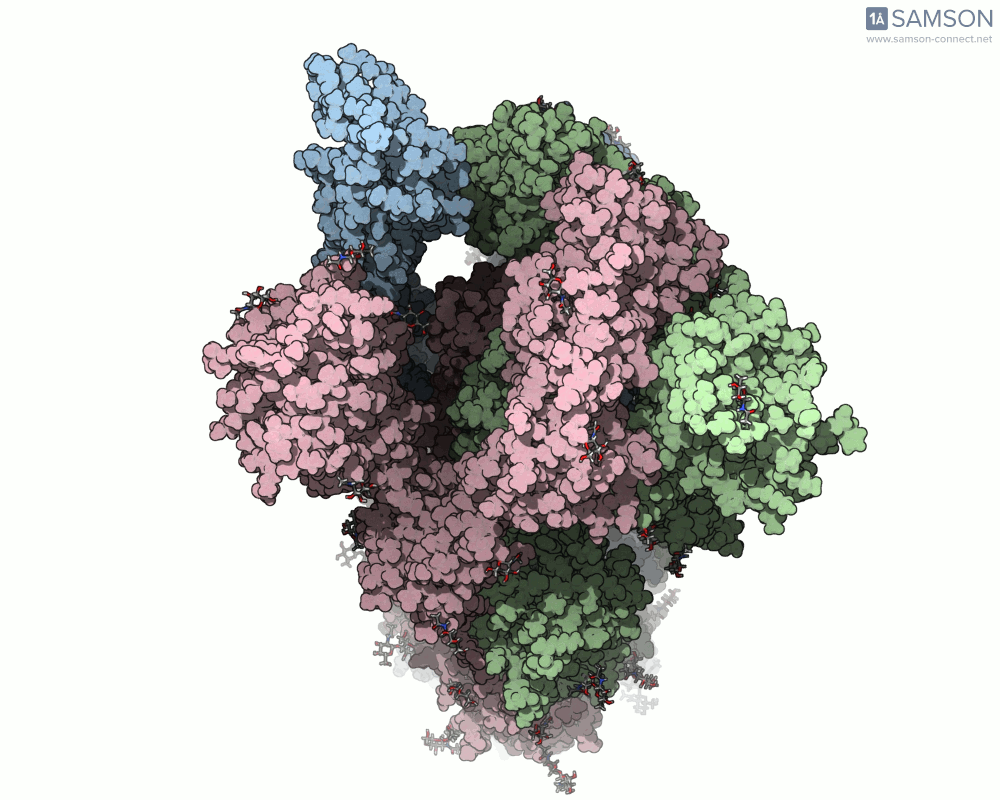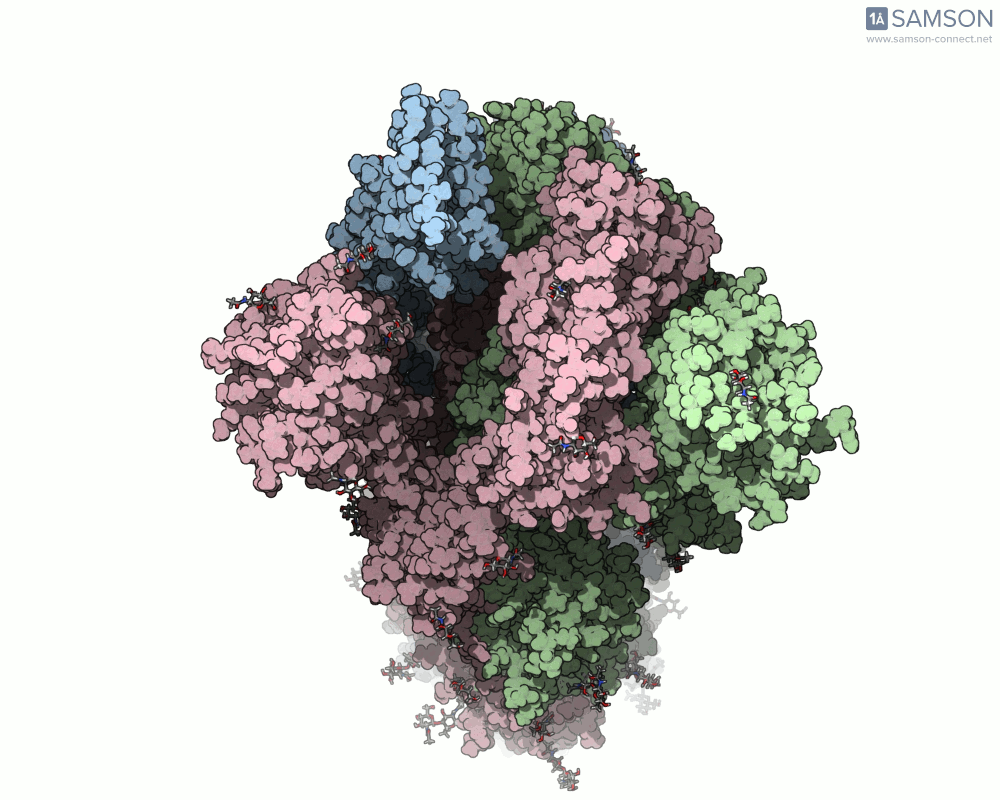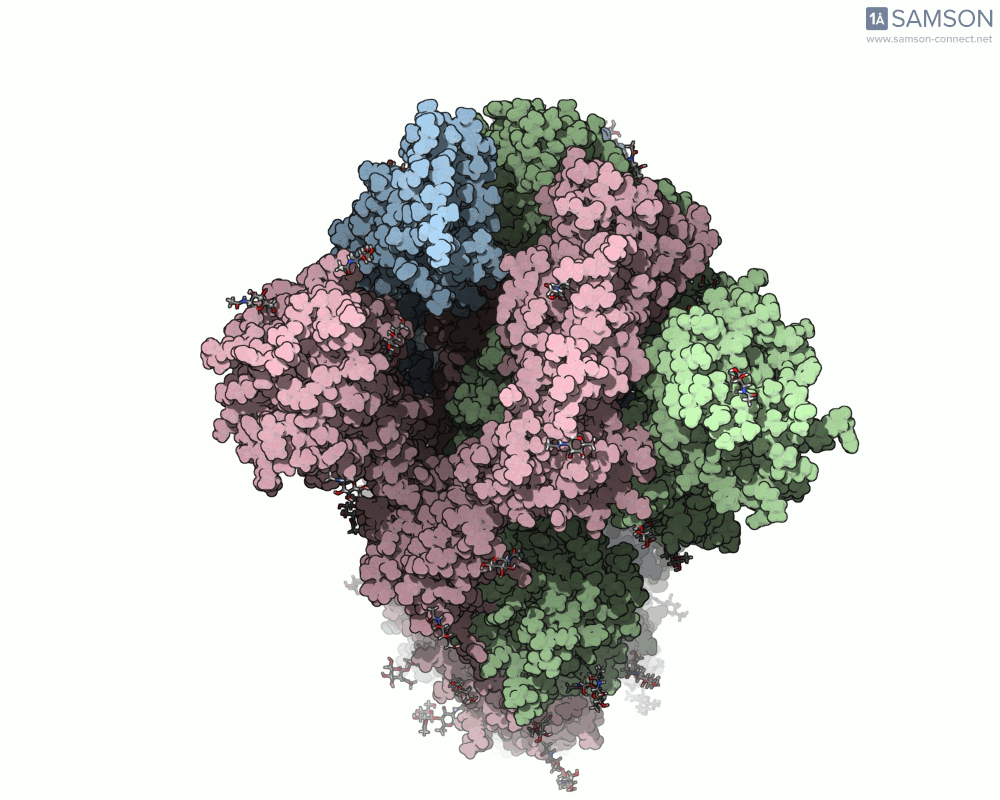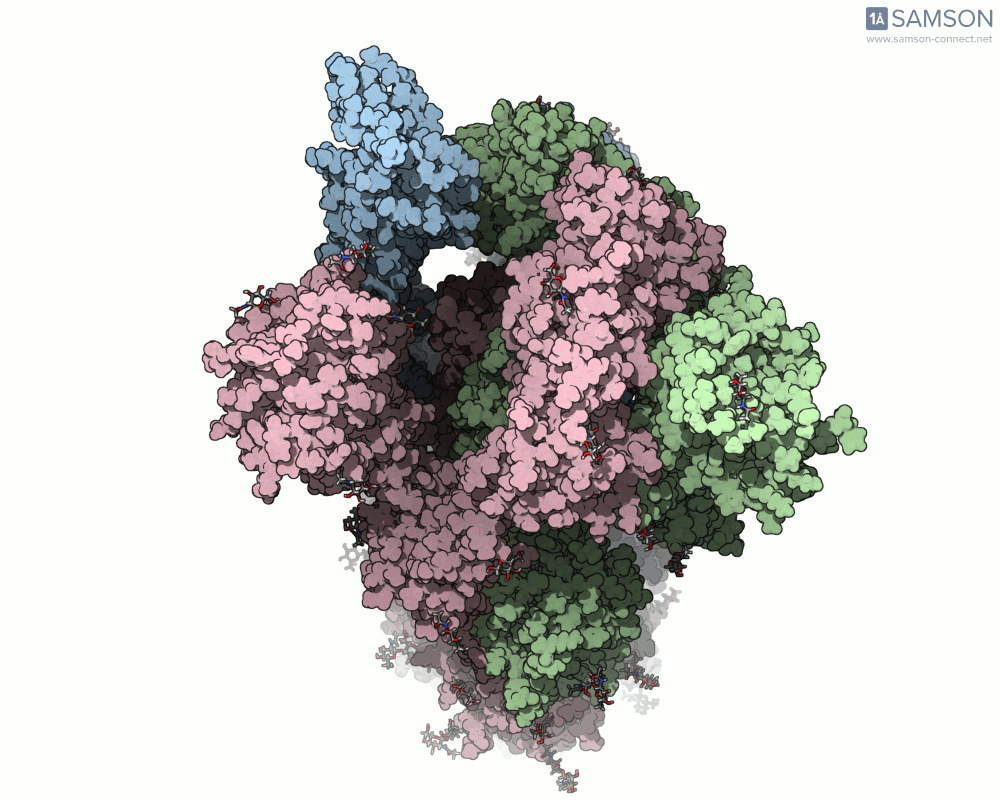
- Top view:
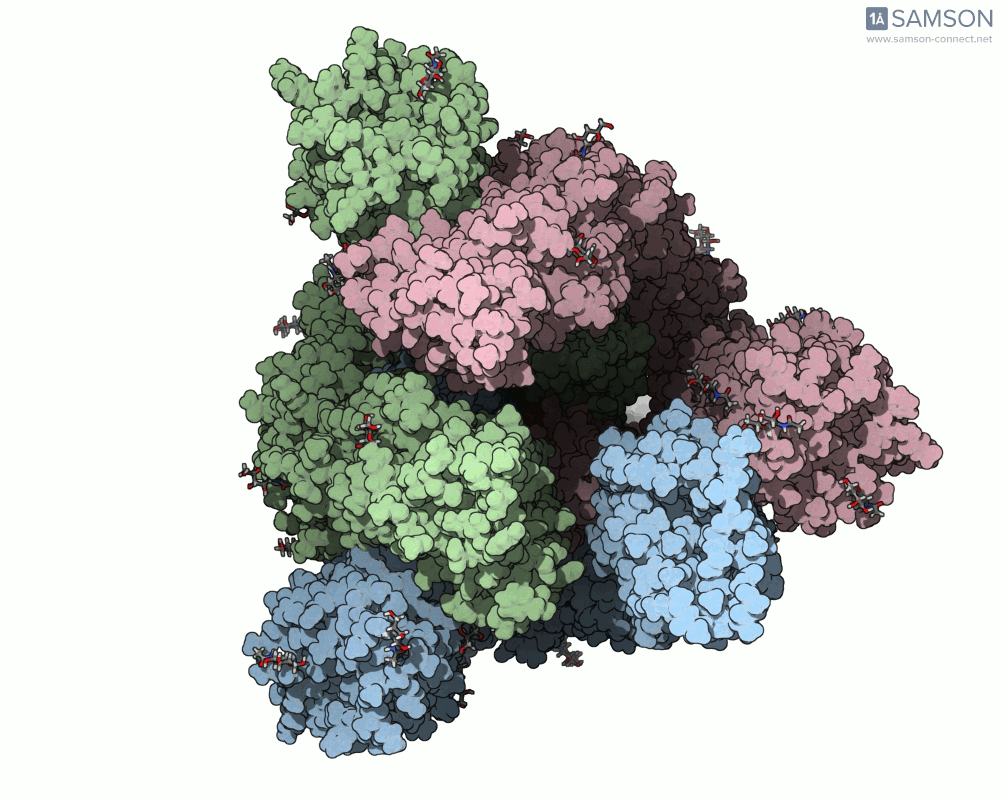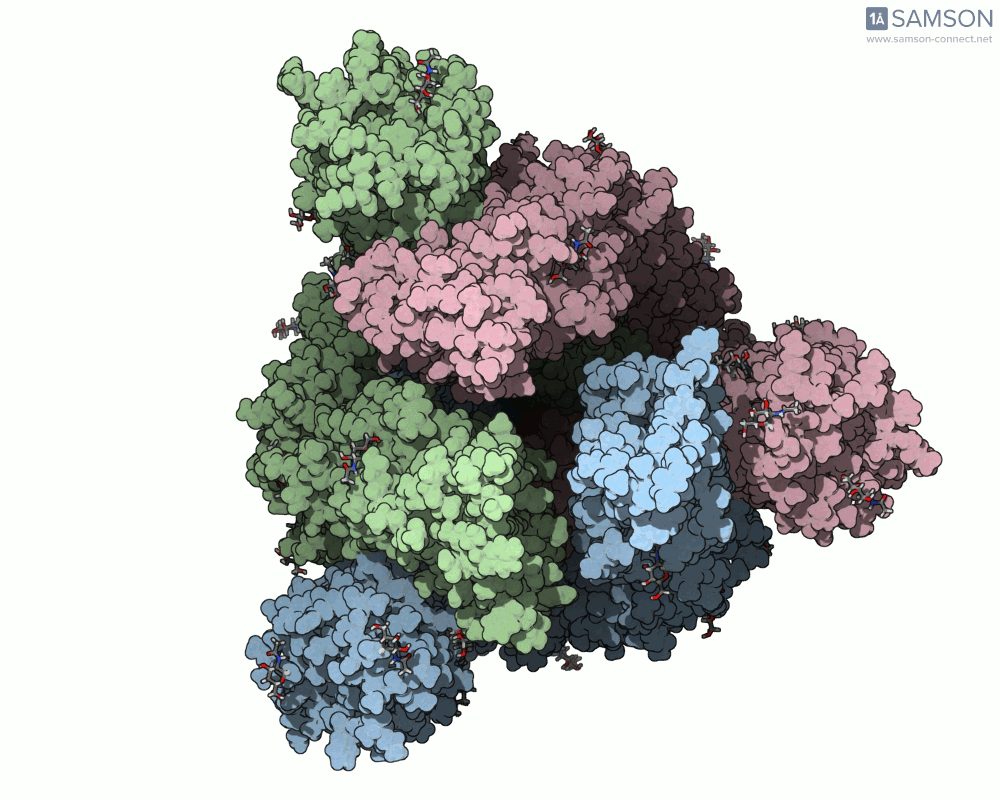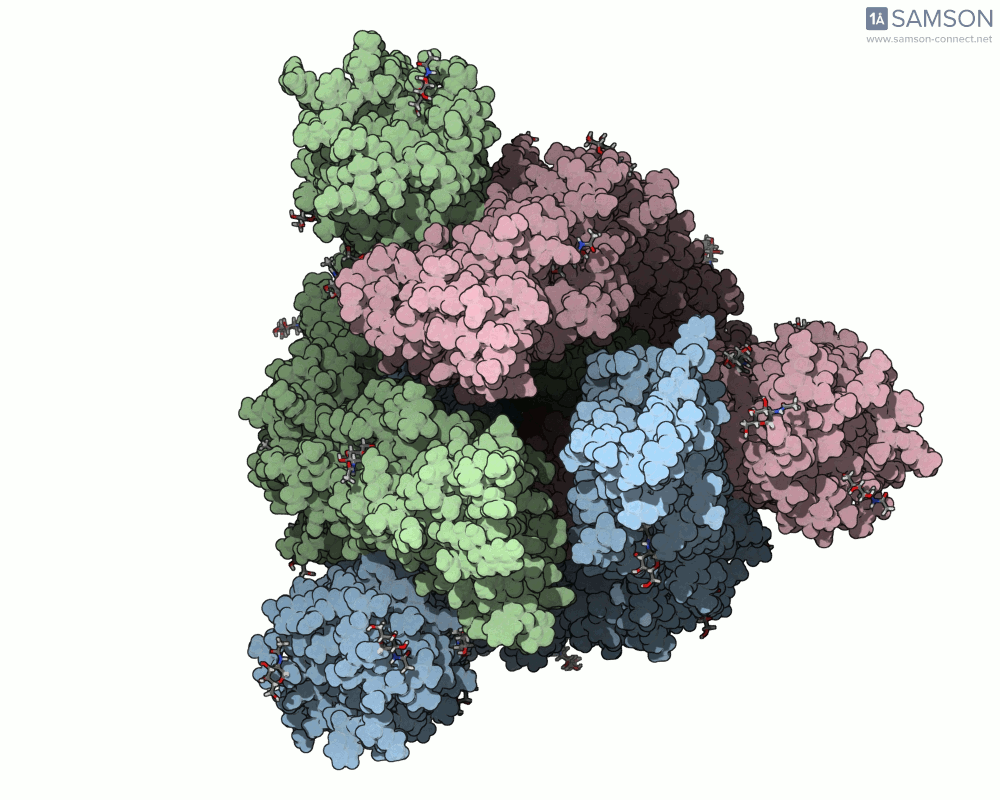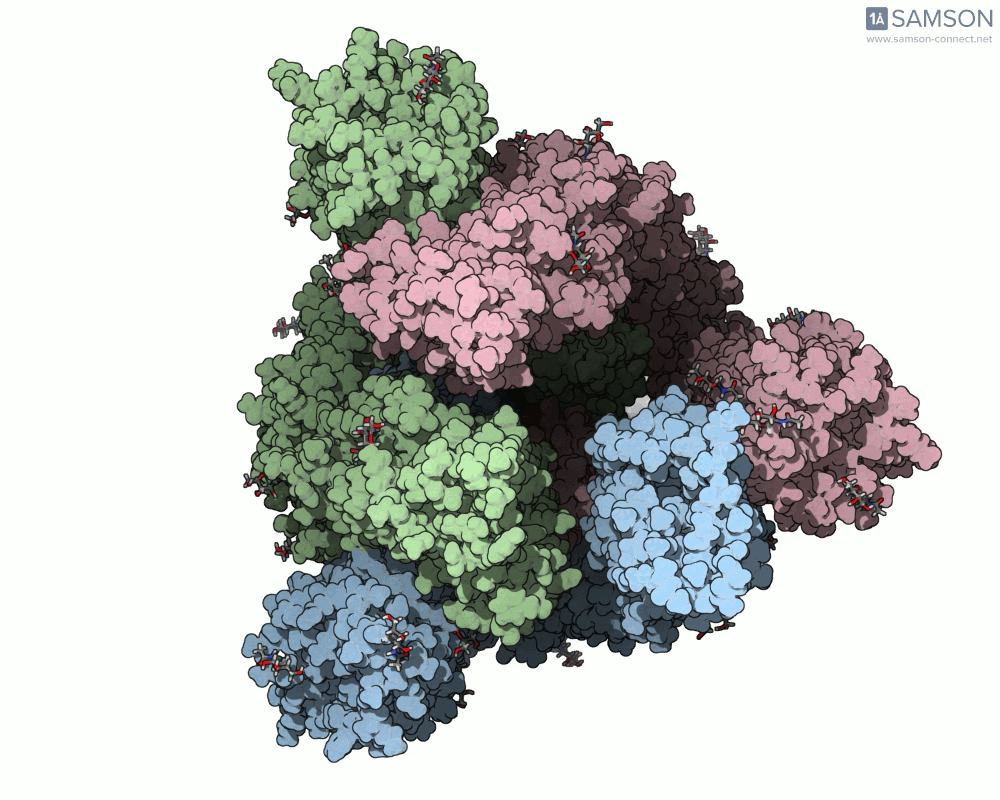
These animations are generated from a computed trajectory using the open (PDB: 6VYB) and closed (PDB: 6VXX) conformations.
Methodology overview
To generate the interpolated trajectory, SAMSON’s developers used a path-computation workflow combining two tools:
- ARAP (As-Rigid-As-Possible) Interpolation: Creates a smooth interpolated path between known conformations by minimizing geometric distortion.
- P-NEB (Parallel Nudged Elastic Band): Refines the path to better represent realistic transitions by minimizing path energy.
This simulation was performed on a laptop in under 20 minutes in total, highlighting that even complex transitions can be computed with accessible resources.
Ready-to-use resources
The computed spike trajectory is shared under a CC BY 4.0 license and available in multiple formats:
These visual resources can serve as inputs for further modeling or teaching workflows, or just as a qualitative tool to better understand viral function. Researchers can customize, analyze, and extend the trajectory right inside SAMSON.
To explore this workflow and download the full example, visit the original documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





