When studying molecular mechanisms such as ligand unbinding, full atomic trajectories are often unnecessary. Many molecular modelers prefer to focus only on a specific subset of atoms—like a ligand or a binding site—when running free energy calculations, visualizing reaction coordinates, or preparing data for enhanced sampling techniques.
The Export Along Paths extension in SAMSON offers a way to export precisely the atomic trajectories you need, avoiding information overload and reducing file sizes. In this post, we’ll walk through how you can select and export only a subset of atoms (e.g., a ligand) along a computed molecular pathway—without the clutter of the rest of the system.
Why Export a Subset?
Let’s say you’ve already generated an unbinding path for a ligand using SAMSON’s Ligand Path Finder. Now, you want to perform a free energy profile by monitoring only the ligand’s movement. Exporting just those coordinates makes your work cleaner, faster, and more focused.
Step-by-Step Guide
Once you’ve loaded your system with a pathway, here’s how you can export a selected group of atoms:
- Open the app via Home > Apps > All > Export Along Paths.
- In the opened interface, expand the Advanced panel.
- In the Document view, select the ligand you want to export (e.g.,
TDG).

- Click Add to define the selection as a model. This registers your selected atoms as a group to be tracked and exported.
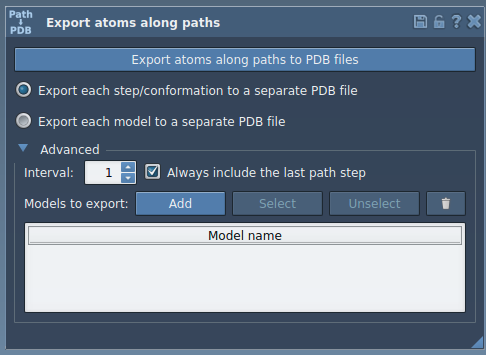
After this, a new row appears in the export table—each row corresponds to a group of atoms you can export:

Customization Options
- Rename the model by double-clicking its name in the table.
- Select/Unselect to visually confirm selected atoms.
- Reset the selection to adjust your atom group.
- Add multiple groups if needed. For instance, track both the ligand and a nearby residue.
Once your subsets are defined:
- Choose your preferred export mode: a single PDB file or one file per frame.
- Select the path(s) your model(s) follow.
- Click Export atoms along paths to PDB files.
This method ensures that only the most relevant parts of your system are saved, streamlining subsequent simulations and analyses.
Common Use Cases
- Export ligand-only coordinates for umbrella sampling setups.
- Generate trajectory snapshots of binding pocket residues only.
- Measure conformational transitions without non-essential atoms.
Whether you’re modeling transport through channels, conformational changes in enzymes, or ligand migration paths, this feature saves time and keeps your data focused—while giving you full control over what’s exported.
To learn more, visit the full documentation page: Export atoms trajectories along paths – Documentation
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON from https://www.samson-connect.net.





