When refining a protein model—whether from homology modeling, docking, or simulation—chances are you’ll encounter a few residues out of place. These are often exposed on a Ramachandran plot as positions falling into disallowed regions, indicating strained backbone conformations. How can you fix these?
The Interactive Ramachandran Plot Extension in SAMSON offers a fast and visual way to address this challenge. With it, you can review, select, and adjust your protein backbone dihedral angles (φ and ψ), while observing real-time changes in 3D. Let’s walk through how to explore and correct strained residues with this approach.
Visually Exploring Residues
Once you load your protein model (say, the structure with PDB ID 1YRF) into SAMSON and open the Ramachandran plot app, the plot displays each residue as a point plotted by its φ and ψ angles.
Color highlights guide you:
- Yellow zones show favorable regions based on established steric constraints
- White zones highlight disallowed conformations
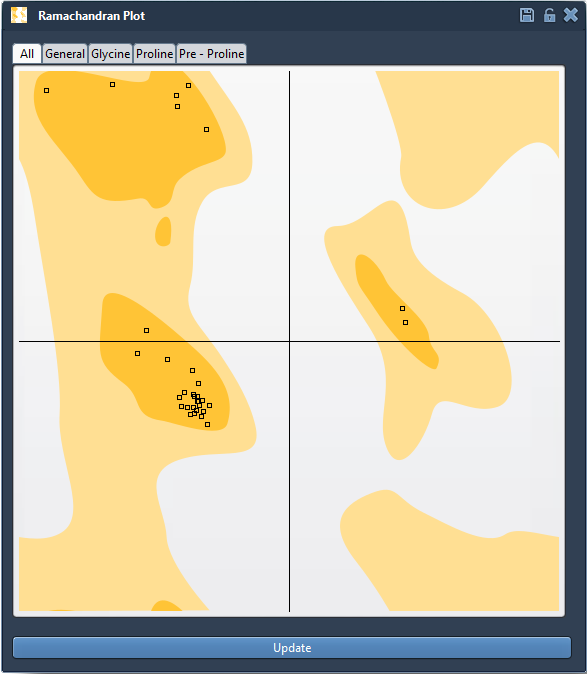
Click any point—like a proline residue—and the corresponding amino acid is immediately selected in the 3D viewport. It also displays its φ and ψ values, helping identify how far a residue has drifted from allowed conformations.
Interactive Correction: Two Editing Modes
1. Dragging Directly in the Plot
- Click and drag a residue’s point on the plot
- Its φ and ψ angles are instantly updated
- The change is reflected in the 3D model in real time
You can always undo with Ctrl / Cmd + Z if something doesn’t look right.
2. Using the Twister Editor
Prefer adjusting the model by hand directly in 3D? Use the Twister Editor:
- Select the Twister icon from the left-hand tools
- Choose your residue
- Click and twist its φ or ψ directly in 3D
- Watch the Ramachandran plot update
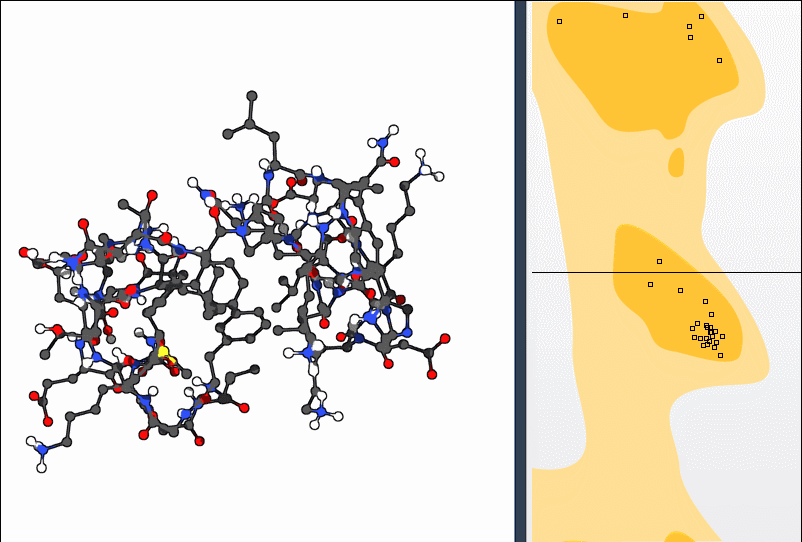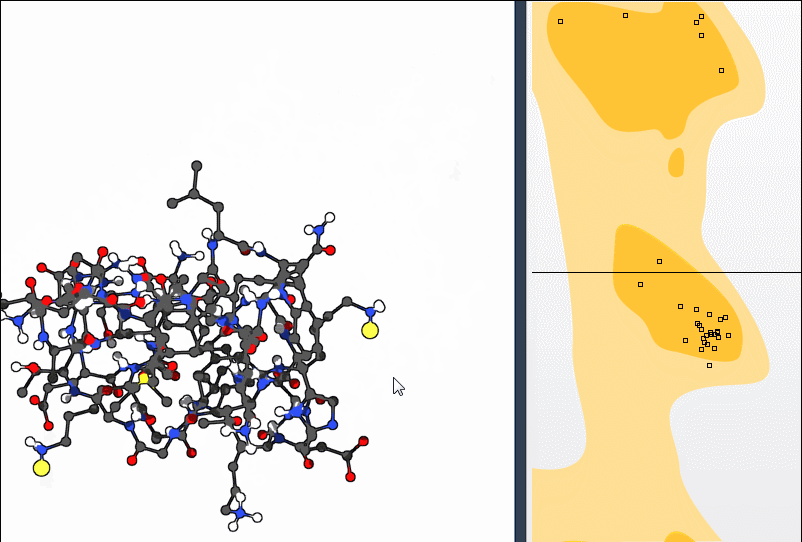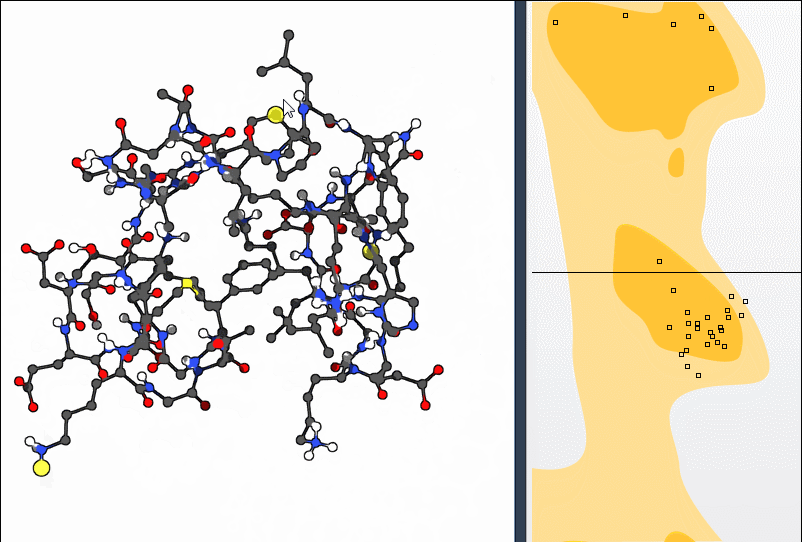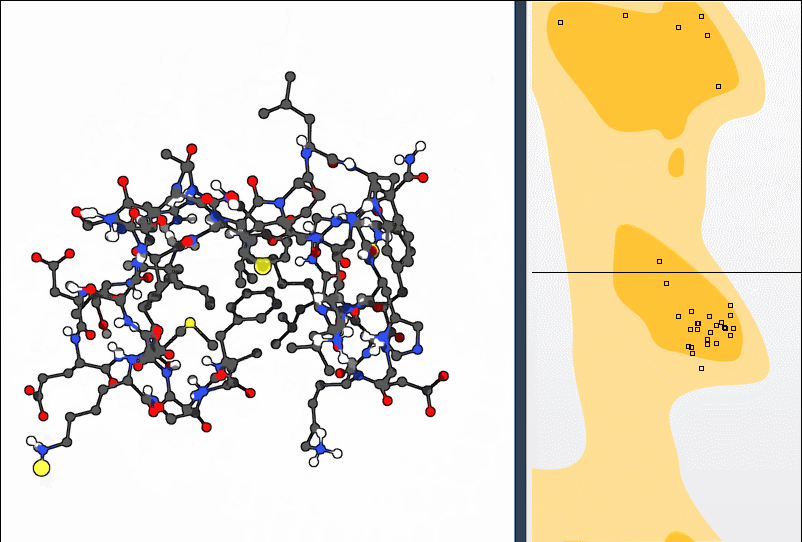
This bidirectional interaction—plot to structure and structure to plot—makes polishing backbone geometry intuitive. You don’t have to guess which atoms to rotate or fiddle with numbers; visual feedback does the work.
Application Tip: Prepare for Simulation
Before launching a molecular dynamics run or energy minimization, it’s smart to correct residues outside favored regions. These often result from model building errors or sidechain clashes and can introduce unrealistic strain into simulations. Even a few outlier residues can disrupt overall structure stability or skew dynamics in early steps.
By interactively dragging them into allowed regions—or making small local adjustments using Twister—you improve the quality of your input structure while maintaining its overall fold. It’s more efficient and visual than rerunning an entire refinement pipeline.
Want to Dive Deeper?
The Interactive Ramachandran Plot Extension integrates seamlessly into biology workflows in SAMSON. For a step-by-step guide with more features, application use-cases, and optional normal-mode based motion refinement, visit the official documentation:
https://documentation.samson-connect.net/tutorials/ramachandran/ramachandran-plot/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





