When studying molecular behavior such as ligand binding and unbinding, free energy calculations, or preparing animations of molecular motion, capturing atom trajectories along computed paths can be crucial. SAMSON’s Export Along Paths extension offers a practical way for molecular modelers to extract atomic coordinates from such paths, particularly when you’re interested in just a few atoms (like a ligand) rather than the entire system.
Consider this: you’ve used SAMSON’s Ligand Path Finder to compute an unbinding path for a ligand. You now want to export only the ligand’s coordinates along this path in a format suitable for downstream simulations, such as umbrella sampling or free energy calculations. Here’s how you can do exactly that using the Export Along Paths extension.
Focusing only on what matters: Exporting a subset of atoms
While you can export all atoms along a path, in many cases you’ll want to export just a subset—say the ligand atoms. This reduces file sizes and allows more focused analysis. Here’s how to isolate what you need:
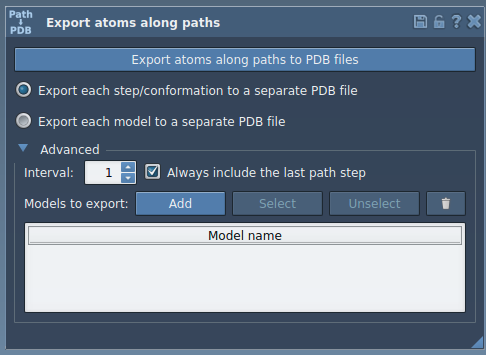
- In the Export Along Paths app interface, expand the Advanced panel.
- In the Document view, select the ligand—in the tutorial example, it’s named
TDG. - Click Add to define this selection as a model to export.
This defines a named group of atoms you’ll export. You can rename the model by double-clicking its name and can repeat the process to add multiple models if needed.
Once satisfied with your selection:
- Choose your export format: a single PDB file or multiple files (one per frame).
- Select the paths along which to export.
- Click Export atoms along paths to PDB files.
You can also adjust how frequently frames are exported by changing the frame interval in the Advanced panel. This is useful if you’re dealing with long trajectories and only need key snapshots.
Why export only part of a system?
Exporting a subset of atoms is especially relevant in scenarios like these:
- Preparing input for reaction coordinate analysis methods (e.g., umbrella sampling).
- Analyzing a ligand’s movement without the distraction of the entire protein environment.
- Creating lightweight trajectory files for visualization or teaching purposes.
By narrowing your focus to just the essential atoms, you make downstream analysis and computation more manageable, faster, and cleaner.
To learn more about exporting atoms along molecular paths, including working with full systems and additional advanced options, visit the full tutorial.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON here.





