Computing the Potential of Mean Force (PMF) is a critical task in molecular modeling, especially when investigating molecular interactions and energetic landscapes. However, running umbrella sampling simulations is only half the job—getting accurate PMF results from the output can be daunting. The GROMACS Wizard in SAMSON makes this easier with integrated support for the Weighted Histogram Analysis Method (WHAM). In this post, we break down how to set up and execute PMF calculations using WHAM in the GROMACS Wizard, helping you turn raw simulation data into insightful free energy profiles.
Why PMF and WHAM Matter
PMF calculations allow molecular modelers to map the free energy profile along a reaction coordinate. This is essential when determining energy barriers, intermediate states, or binding affinities. WHAM is widely used for combining multiple biased simulations (like umbrella sampling windows) into a statistically rigorous PMF curve. Yet, parsing output files, configuring bounds, and scripting data analysis isn’t for everyone. That’s why having an integrated tool like GROMACS Wizard can be a time-saver.
Step-by-Step: Using WHAM in GROMACS Wizard
After completing your umbrella sampling simulations—which ideally were run with the help of GROMACS Wizard’s batch computation feature—you can switch to the WHAM Analysis tab to process the data. Here’s how:
- Open GROMACS Wizard in SAMSON and go to the WHAM Analysis tab.
- Select your project path. If you’ve used the previous steps in the Wizard, click the auto-fill button
 to import your last used path automatically.
to import your last used path automatically. - Make sure your folder contains numbered subdirectories (e.g.,
000,001, …) representing different windows from the umbrella sampling. Each should include simulation results for the same reaction coordinate and settings.
- The Wizard will extract relevant data such as reaction coordinate names, bounds, time, and temperature.
- Choose your desired reaction coordinate from the list.
- Configure options like custom bounds, time intervals, and energy units if needed.
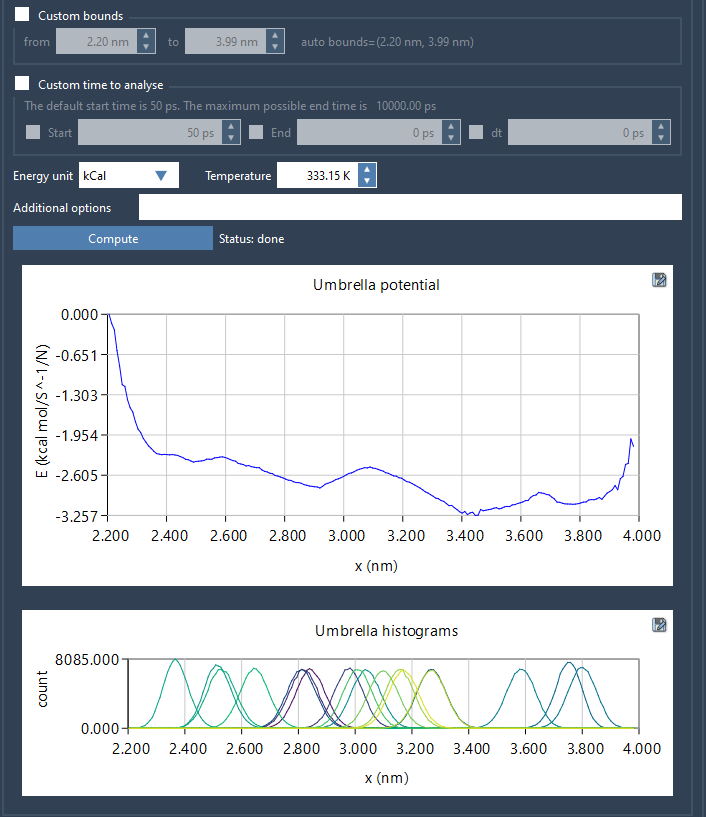
- Click Compute. This will launch the WHAM calculation and generate two key plots:
- PMF curve: Gives the free energy profile across your reaction coordinate.
- Histogram: Shows sampling density and helps identify regions under-sampled during umbrella sampling.
Tips and Benefits
Once the computation is done, results are saved in the wham_results subfolder, making it easy to revisit or change parameters without rerunning simulations. Switching between different reaction coordinates will automatically use cached data where applicable.
This integration eliminates manual steps, reduces setup errors, and offers clear visual feedback on the quality of sampling and the resulting energy profile. Whether you’re prototyping a binding event or refining a pathway study, this workflow focuses your attention on interpreting results, not wrestling with command-line intricacies.
To learn more, see the full documentation on PMF analysis in GROMACS Wizard.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON here.





