Creating realistic animations of conformational changes in biomolecules often feels like a daunting challenge. Full molecular dynamics (MD) simulations can take days or even weeks to run, and demand high-performance computing resources. But what if you simply want to visually interpolate between two known conformations of a protein—say, the open and closed states of the SARS-CoV-2 spike—to understand a mechanism or prepare a presentation?
That’s a common pain point for many molecular modelers: when simulations aren’t feasible or necessary, but a trajectory is still needed. A simple, fast solution? Try trajectory interpolation based on geometry-aware algorithms like ARAP and P-NEB in SAMSON.
From static to dynamic: Interpolating spike protein motions
The SAMSON team has demonstrated how to use the platform to compute the transition from the closed to open conformation of the SARS-CoV-2 spike protein. Using structures from the Protein Data Bank (6VXX for closed, 6VYB for open), they generated a smooth transition without running MD. Here’s how they did it:
- Pre-processing: Added hydrogens and fixed bond orders on sugars via a Python script.
- Initial interpolation: Used the ARAP (As-Rigid-As-Possible) module to quickly interpolate from open to closed state.
- Post-processing: Refined the path with the P-NEB (Parallel Nudged Elastic Band) module for better energetic plausibility.
The result is a visually informative and structurally plausible trajectory. It’s not an MD simulation, but it generates transitions in seconds and can be fine-tuned if needed. Check out how the spike protein moves from closed to open in the GIF below:
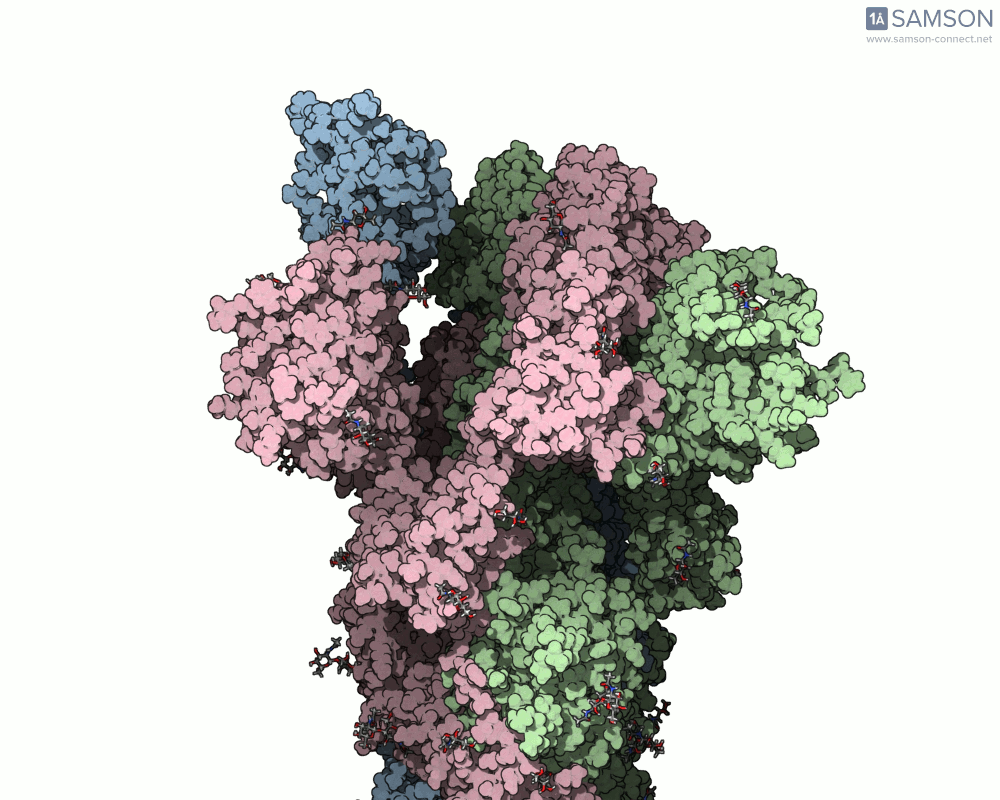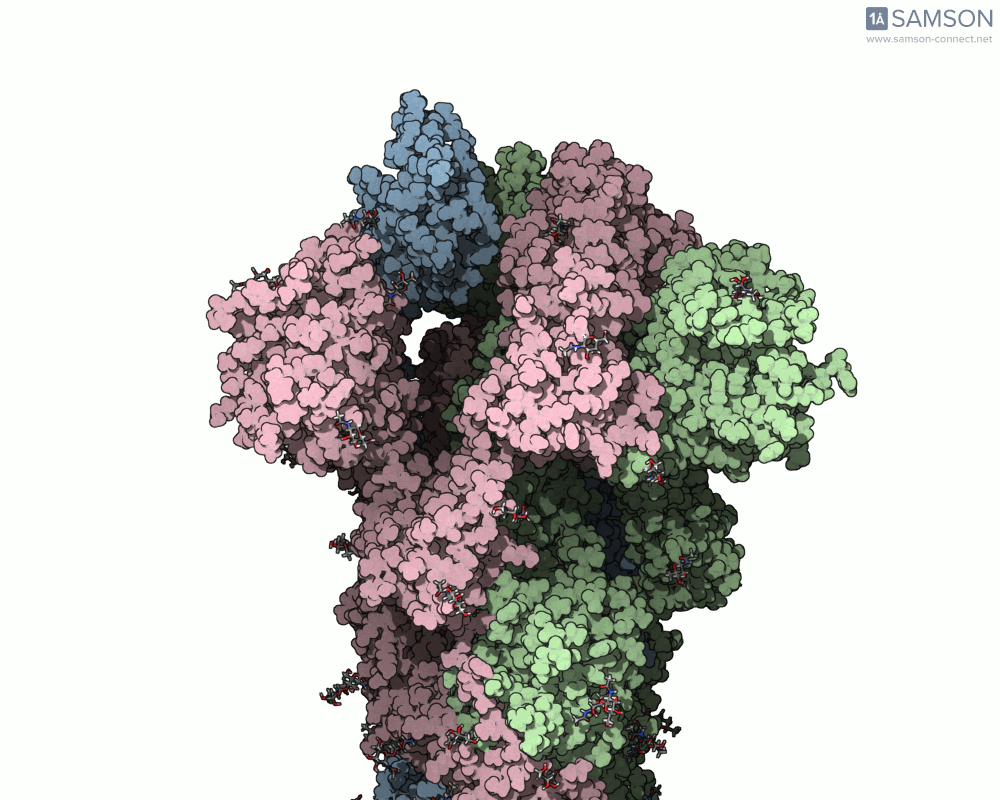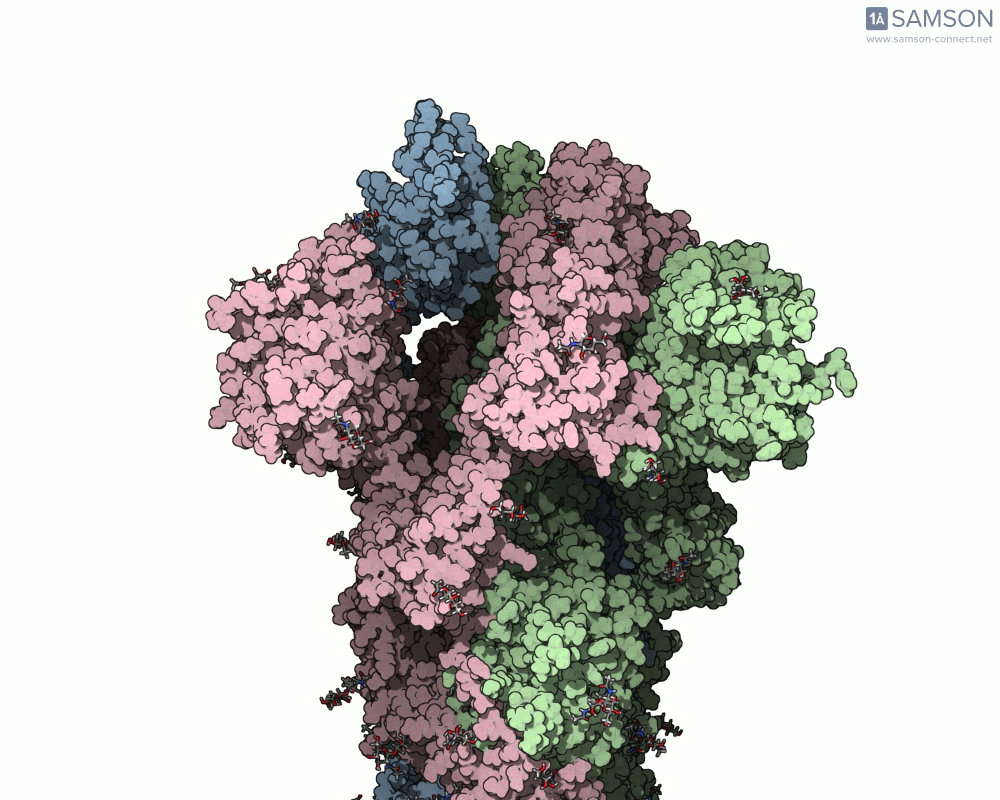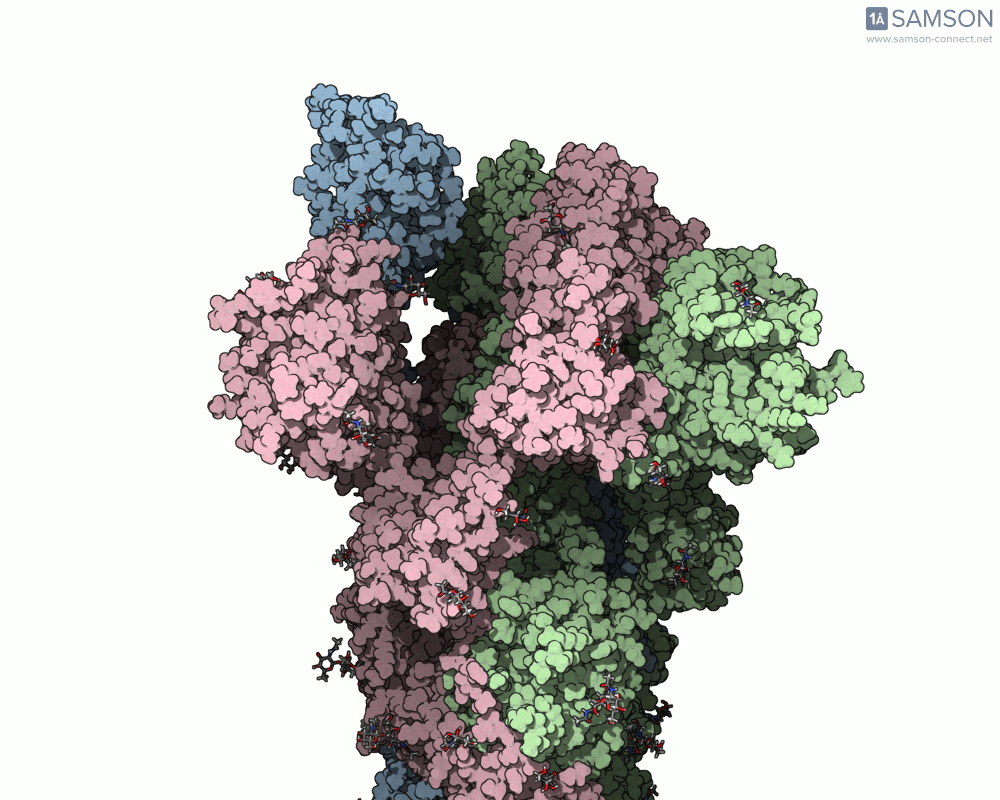
When should you use this approach?
This method is ideal when:
- You have two known conformations and need an illustrative transition.
- You want quick insights into possible movement mechanisms.
- You’re preparing educational or scientific visuals and don’t have time (or compute resources) to run MD.
- You want to initialize more refined simulations using a plausible intermediate state.
Export and use the results
The computed trajectory is directly available in several formats:
These outputs can be loaded in visualization tools or reused as part of other simulations. The SAMSON document even contains trackable conformations and animations.
To learn more about this workflow and see additional animations, visit the original documentation page:
https://documentation.samson-connect.net/tutorials/sars-cov-2/coronavirus-computing-the-opening-motion-of-the-sars-cov-2-spike/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





