Preparing ligands for docking can often slow down workflows in molecular modeling. Whether you’re running structure-based virtual screening campaigns or docking a few analogs to evaluate potential binding modes, the quality of your ligand structures directly impacts the reliability of your docking results. But what if you could streamline ligand preparation into a few clicks? In this post, we focus on using the AutoDock Vina Extended extension in SAMSON to quickly get your ligands ready for docking, minimizing manual processing.
Why Ligand Preparation Matters
Docking algorithms like AutoDock Vina require properly prepared ligands to function accurately: correct hydrogen placement, reasonable 3D conformations, and well-defined rotatable bonds. Skipping this step can lead to poor scoring, unrealistic poses, or failed computations.
Automated Ligand Minimization and Hydrogen Addition
Using SAMSON’s AutoDock Vina Extended app, you can automatically minimize ligands and add missing hydrogens before docking. This is especially useful when working with large ligand libraries, or when your input contains 2D structures or incomplete 3D files.
To access the minimization feature:
- Load your ligand library into AutoDock Vina Extended by checking the Ligand library option and browsing for your ligand files.
- Select the Minimize checkbox to trigger automatic minimization.
- Choose a minimization preset — default options control the number of steps and stopping criteria.
- If your molecules lack hydrogens, check Add missing hydrogens. This is essential for correct bond typing and for identifying hydrogen bond donors/acceptors during docking.

Controlling Ligand Flexibility
By default, ligands are treated as flexible, which usually increases docking accuracy but also computational time. To fine-tune this:
- Use the Lock specific ligand bonds option if you want to make certain bond types non-rotatable during the docking step.
- Click on Locked bonds settings to choose which bond types (e.g., amide, peptide) to exclude from rotation.
Here’s how to do it visually:
- Once a ligand is added, interactive green and red bond controllers appear in the Viewport.
- Click on a controller to toggle a bond’s flexibility. Green means rotatable; red means fixed.
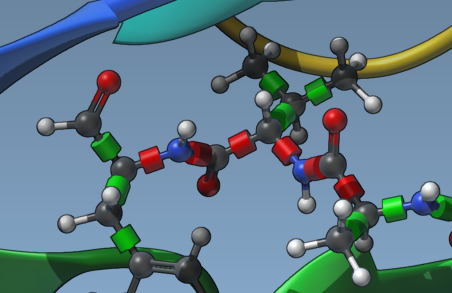
These controls give you per-bond flexibility settings without needing to edit files manually.
Best Practices
- Always run minimization if your ligands come from 2D sources like SMILES or have inconsistent geometry.
- Ensure hydrogens are added before docking to capture interactions accurately.
- Pre-check rotatable bonds for unwanted flexibility in rings or amide bonds.
Conclusion
Proper ligand preparation doesn’t have to involve jumping between multiple tools. Using AutoDock Vina Extended in SAMSON, you can minimize and preprocess your ligands directly inside the docking workflow, saving time and reducing errors — especially helpful for screening large libraries.
To learn more about ligand preparation and docking workflows in SAMSON, visit the full tutorial at this documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON here.





