When working with protein structures from the Protein Data Bank (PDB), researchers commonly analyze individual asymmetric units. However, much of the biological function and structural insight lies beyond the asymmetric unit — in the full biological assembly. These complete assemblies provide critical insights into protein-protein interfaces, quaternary structures, and sites of molecular interaction. But how can you reconstruct these assemblies from a single PDB file?
That’s where the Symmetry Mate Editor in SAMSON can help. This extension rebuilds symmetric replicas of proteins using the transformation records (CRYST1 and BIOMT) stored inside PDB files. The result is a more complete picture of your protein — one that can be used in docking, simulation, and design workflows.
Quickly Visualize Crystal and Biological Symmetries
The Symmetry Mate Editor leverages two types of PDB symmetry data:
- CRYST1 records: Describe crystal lattice symmetry.
- BIOMT records: Encode biological assembly symmetries suggested by experimental authors.
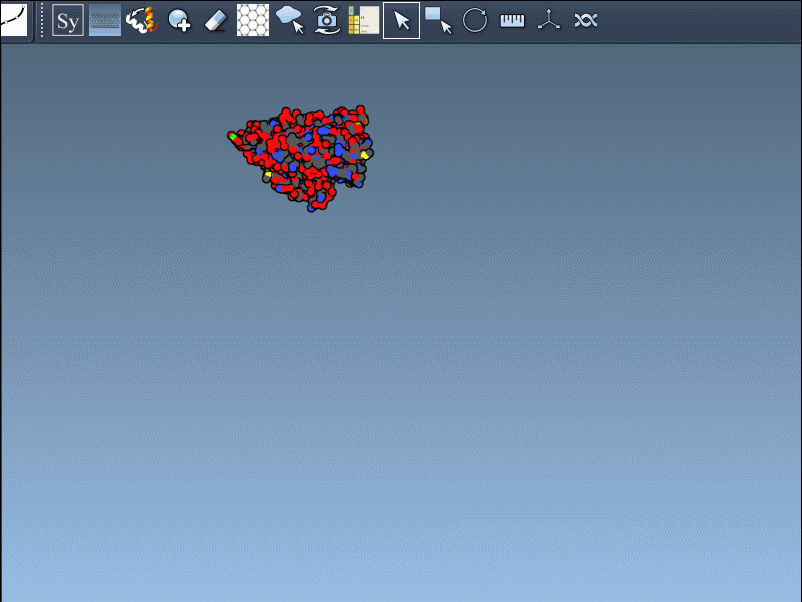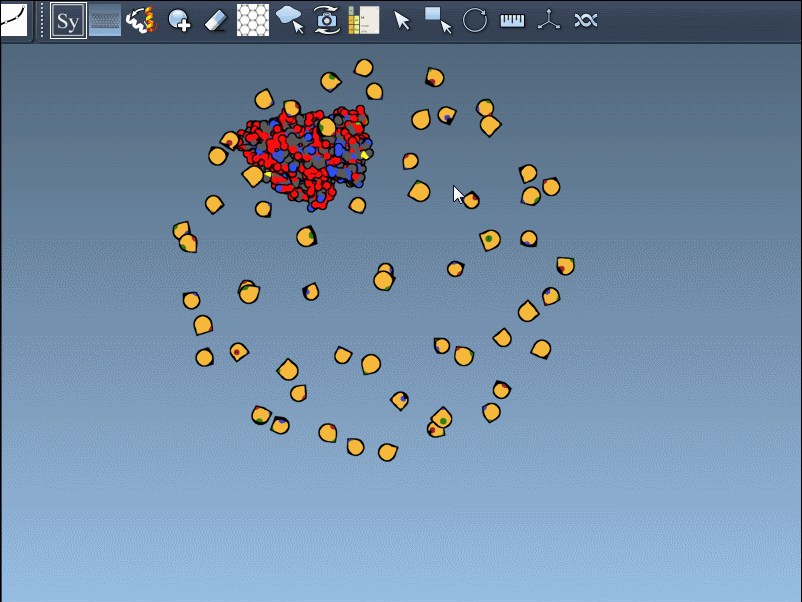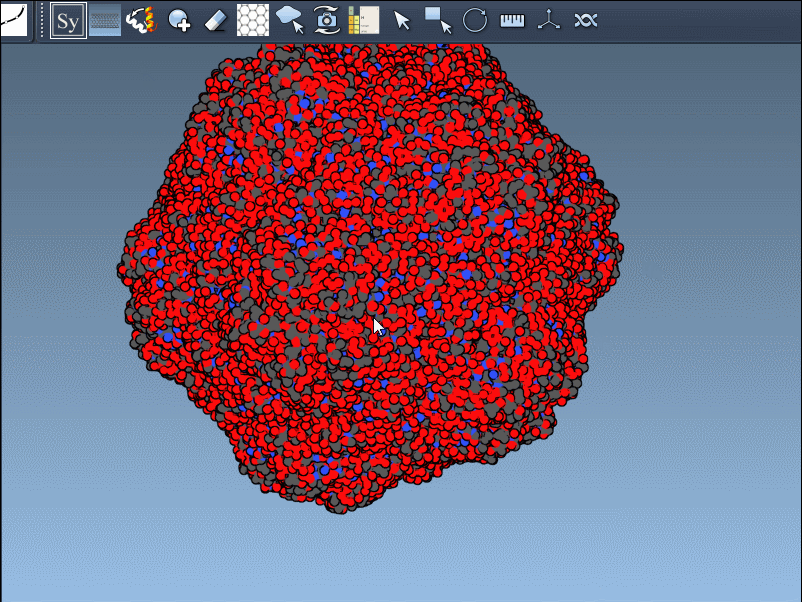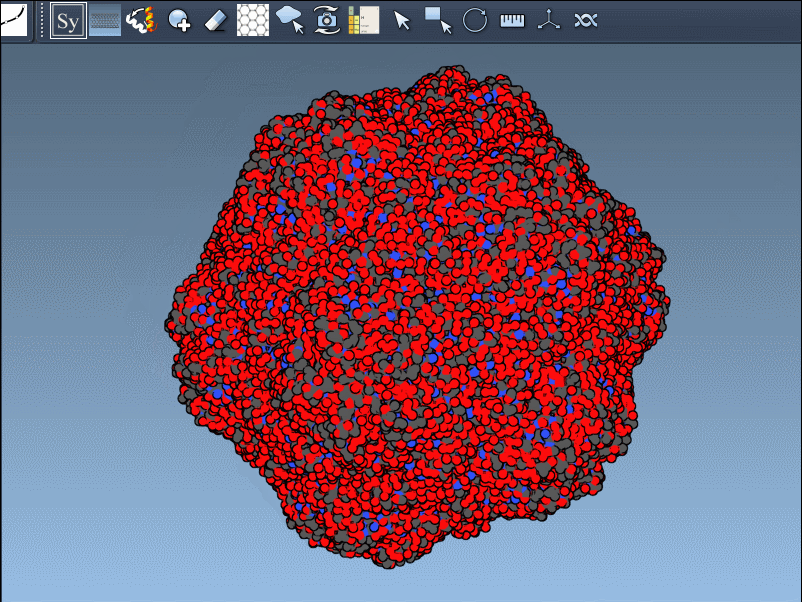
These symmetries are visually indicated in SAMSON using control nodes — white for CRYST1 and yellow for BIOMT transformations:
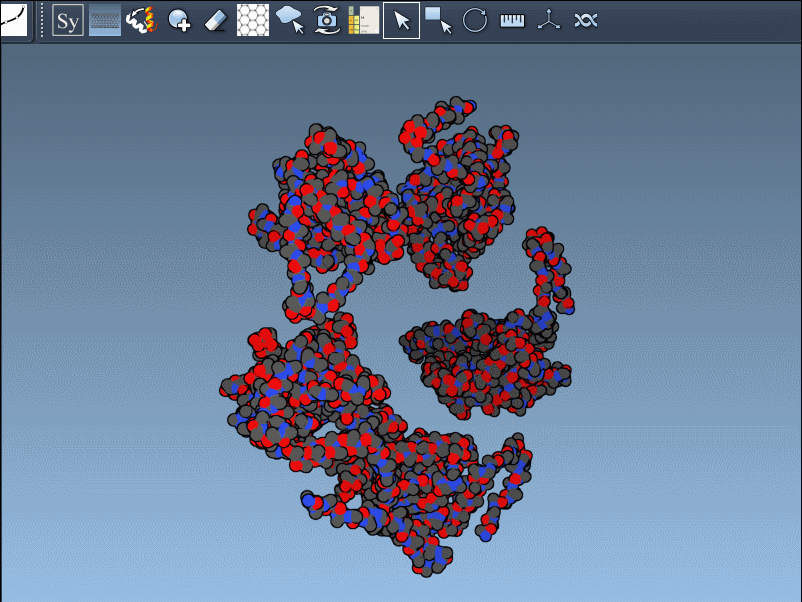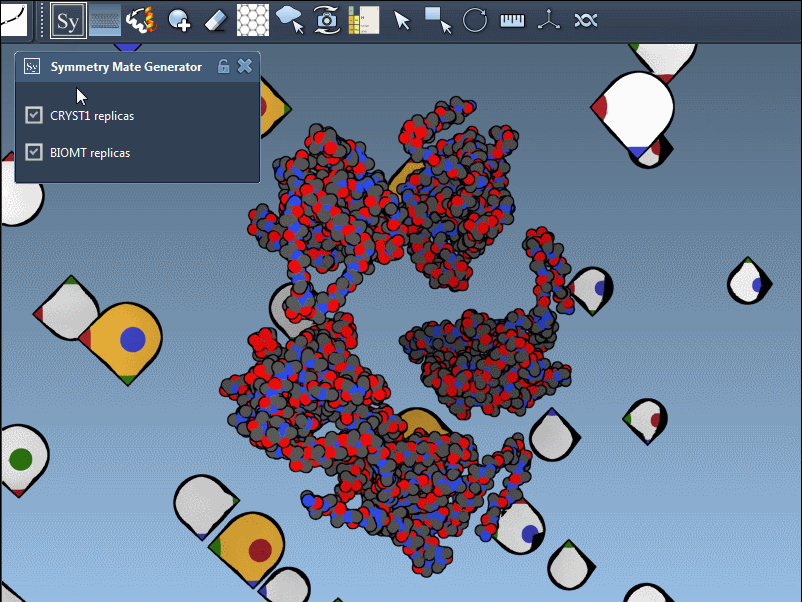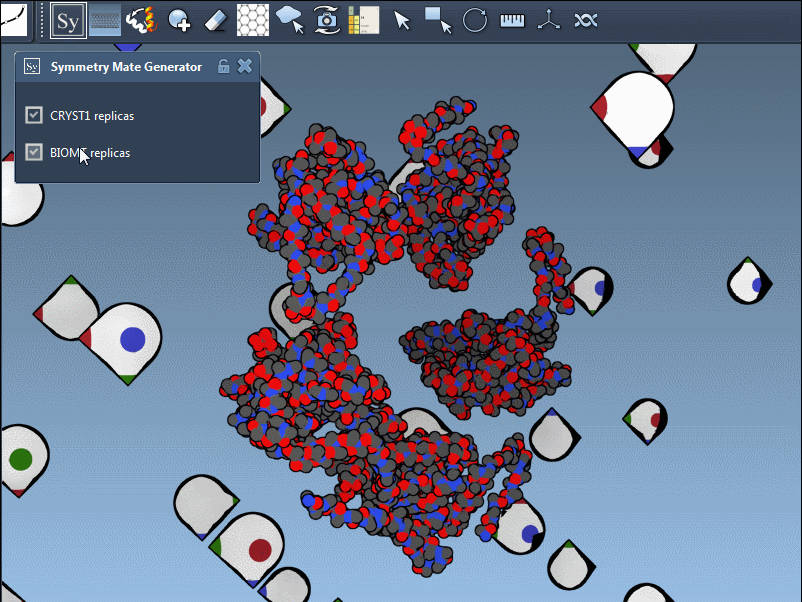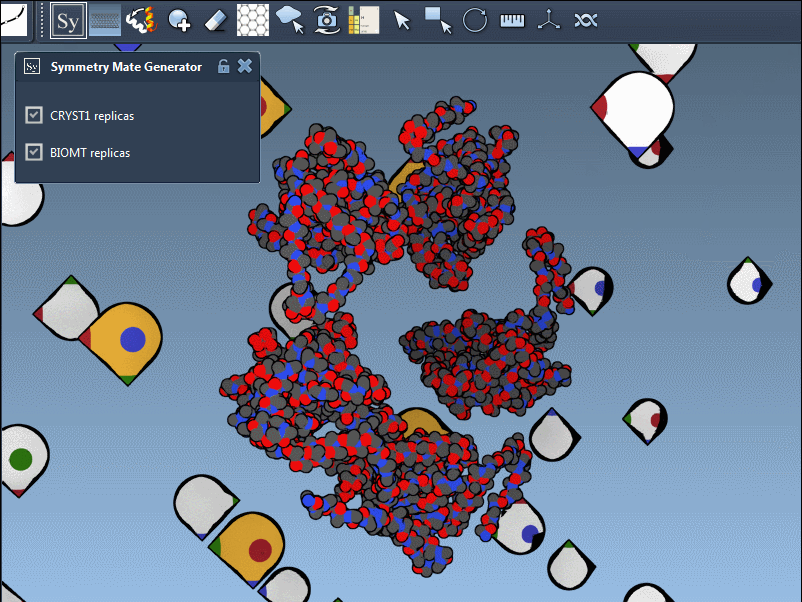
Hovering over a node previews the corresponding replica, and clicking it generates a permanent copy. This interactive workflow helps researchers better understand how residues interface across symmetry mates or find repeating motifs useful in supramolecular design.
From Asymmetric Unit to Full Structure in Seconds
Generating all replicas at once is also straightforward. Just press and hold Ctrl (Windows/Linux) or Cmd (macOS) while hovering and left-clicking a node. All symmetry mates defined by that transformation set will be generated instantly:
This feature is particularly useful when working with large oligomeric assemblies or symmetric cages, where manual generation of multiple replicas would be time-consuming.
Applications in Molecular Modeling
- Dimerization and multimerization studies: Examine protein interfaces by reconstructing dimers, tetramers, or larger assemblies from monomer data.
- Symmetric docking: Use repeated structures to infer potential multivalent binding interactions.
- Molecular dynamics preparation: Create a full assembly before energy minimization, equilibration, or simulation.
Visualizing the replicated structure in SAMSON can also help you determine whether interactions observed in crystallography are biologically relevant or crystallographic artifacts.
Enhance Clarity with Visualization Tools
To make symmetry mates easier to interpret:
- Activate the Ribbons visual model and assign different chain colors.
- Undo misplaced replicas with Ctrl/Cmd + Z.
Conclusion
Understanding symmetry in protein structures is essential for anyone modeling macromolecular assemblies. Whether you’re designing protein cages, analyzing structural interfaces, or preparing biological assemblies for simulation, the Symmetry Mate Editor helps streamline your workflow and reduces manual reconstruction efforts.
To explore more, visit the full documentation here: https://documentation.samson-connect.net/tutorials/symmetry/generating-symmetry-mates/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





