Setting up realistic molecular environments can be a daunting task for molecular modelers. One recurring challenge is positioning lipids around transmembrane proteins to initiate simulations that mimic biological membranes. Manually assembling lipid bilayers or layers is not only time-consuming but also error-prone.
Fortunately, the Molecular Box Builder extension for SAMSON offers a straightforward way to populate a 3D box with molecules — including lipids around proteins — with a few guided steps. In this post, we walk you through creating a lipid bilayer around a membrane protein using this feature. This could be especially useful before starting molecular dynamics simulations or docking studies involving membrane-bound systems.
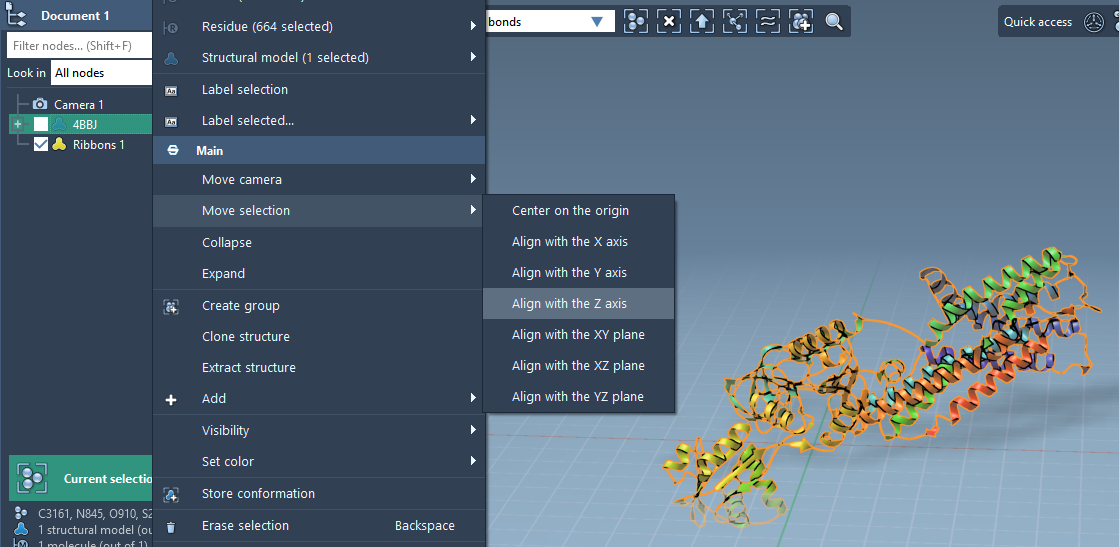
Step 1: Align the Protein
To ensure correct bilayer orientation, start by aligning your protein of interest along the Z-axis. For instance, if you are working with the membrane protein 4BBJ:
- Open the structure in SAMSON (e.g., directly from RCSB PDB).
- Right-click the protein in the Document view.
- Choose Move Selection > Align with Z axis.
- Next, Move Selection > Center on the origin.
Step 2: Set the Lipid Molecule
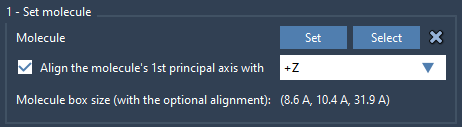
After loading the lipid structure (e.g., POPC), select it in the document and click Set in the Molecular Box Builder app. To ensure the lipid tails align properly against the membrane plane, align the lipid’s principal axis to the +Z direction.
Step 3: Define the Box
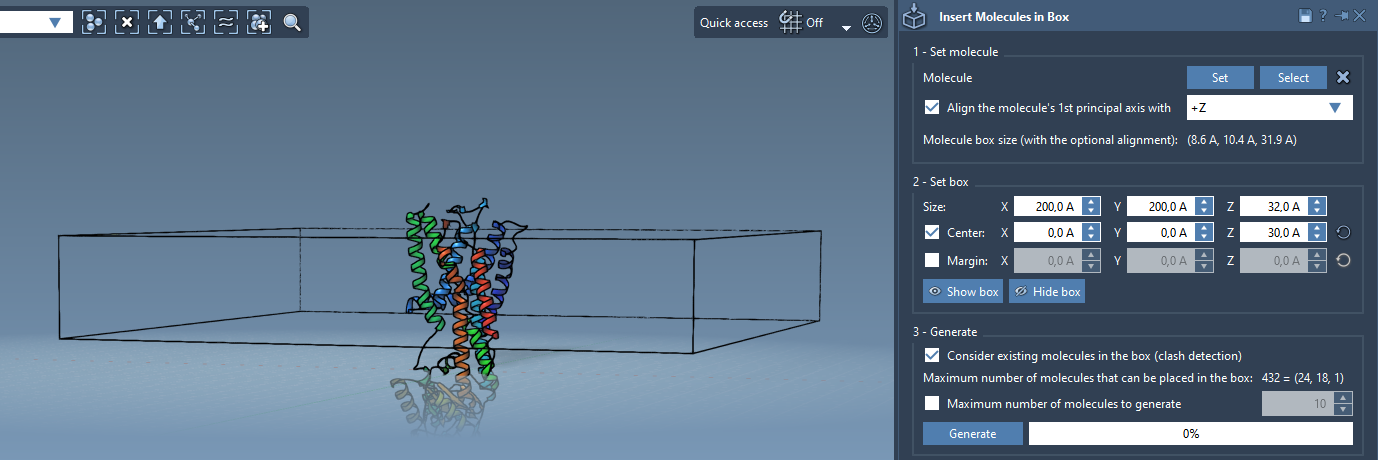
The next step is defining the box where lipids will be inserted to surround the protein:
- Center the box on the protein structure.
- Adjust the box dimensions to encompass the transmembrane portion, leaving room for a single lipid layer.
- Specify a margin between inserted molecules if needed, typically to prevent overlaps.
Step 4: Generate the System
You can now populate the box with lipid molecules:
- Activate Consider existing molecules in the box to prevent lipids from clashing with the protein.
- Click Generate.
The result is a neatly packed lipid layer surrounding the protein.

Optional: Create a Bilayer
To build both upper and lower membrane leaflets:
- Generate the first lipid layer with
+Zalignment. - Shift the box slightly along the
Zaxis to position the second layer below the first. - Generate another layer using the same lipid structure, this time aligned with
-Z.
This gives you a full lipid bilayer enveloping your membrane protein—a useful setup for simulations such as membrane insertion, receptor-ligand studies, and more.
Learn more at the full documentation page: Molecular Box Builder Documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON from https://www.samson-connect.net.





