For many molecular modelers, building lipid membranes around membrane proteins still requires tedious setup or reliance on external, specialized tools. Whether you’re preparing a simulation system for dynamics or modeling a membrane-embedded protein, ensuring that your lipid bilayer is correctly positioned and packed can be surprisingly time-consuming.
The Molecular Box Builder extension in SAMSON helps simplify this process. In this post, we’ll walk through a practical example: how to build a lipid layer (or bilayer) around a membrane protein using Molecular Box Builder’s intuitive interface. No scripting, no external tools—just an integrated, visual approach that fits directly into your SAMSON workflow.
Why this matters
If you’ve ever tried packing lipids around a protein, you know it’s about more than just space—it’s about alignment, exclusion of overlapping molecules, and respecting system symmetry. Incorrect orientation leads to unrealistic structures, and poorly spaced lipids can even crash simulations. Let’s take a closer look at how to avoid these issues from within SAMSON.
Step 1: Align the Protein
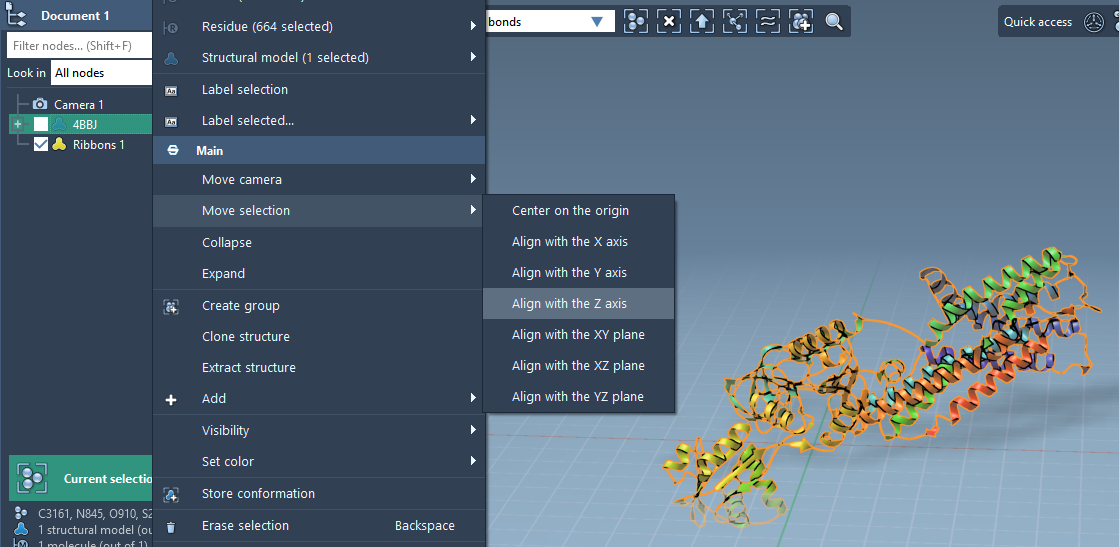
Start by orienting your membrane protein (e.g., 4BBJ) with its membrane-spanning region along the Z-axis. This sets up a natural axis for placing lipids along the membrane plane.
- In the Document view, right-click your protein
- Select Move selection > Align with Z axis
- Then choose Move selection > Center on the origin
Step 2: Select and Align the Lipid
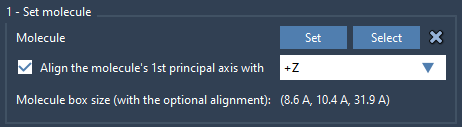
Import your lipid molecule (e.g., POPC), then align it along the +Z direction. This ensures the lipid tails point in the correct direction when added to the membrane.
- Import the lipid into SAMSON
- Select it and click Set in the Molecular Box Builder app
- Choose to align it along
+Z
Step 3: Define the Box
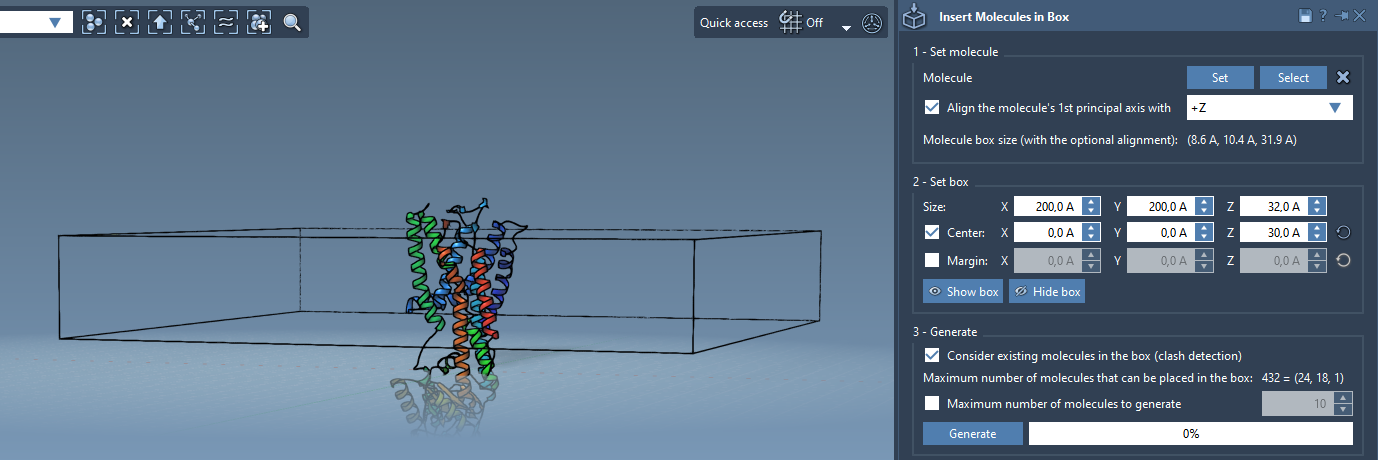
Next, center the box around the protein. Adjust the box’s dimensions (especially along Z) to allow for one or more lipid layers. The side margins help control how tightly lipids are packed.
- Tune X and Y to surround the protein’s footprint
- Adjust Z for a single or double lipid layer (for bilayers)
- Use the Center checkbox to anchor the box properly
Step 4: Generate the Lipid Layer
To avoid lipid overlaps with your protein, activate the checkbox Consider existing molecules in the box. Then simply click Generate. Lipids will automatically be placed only in unoccupied space.

Want a full bilayer?
Here’s how to build it:
- Generate the first layer with
+Zalignment - Shift the box down along Z
- Regenerate a second layer with
-Zalignment
This sequence creates a bilayer with lipids in both leaflets oriented correctly.
Final thoughts
Simple steps and visual feedback take away the uncertainty of lipid packing around membrane proteins. Because it’s tightly integrated into SAMSON, you can immediately proceed to system minimization or export to MD simulation tools.
Learn more in the full documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON here.





