One of the most common scenarios in molecular modeling is embedding a membrane protein into a lipid bilayer. While it sounds straightforward, anyone who has tried to manually position hundreds of lipids around a protein knows this can be a time-consuming task prone to errors and steric clashes.
Fortunately, the Molecular Box Builder in SAMSON simplifies this process. With just a few clicks, you can generate a bilayer of lipids aligned to a desired orientation around your protein – without interpenetrating molecules. Here’s how it works, and why it saves so much time.
Why Build the Membrane This Way?
Manually constructing lipid environments often involves downloading pre-equilibrated membranes, aligning them with the protein, trimming or adding lipids, and checking for overlaps. The Molecular Box Builder turns this into a more deterministic process. Instead of random placement, it arranges molecules in tightly packed grids inside a box. When that box is defined around a protein, it respects the occupied space and fills only the empty regions, making clash-free insertion easy.
Step-by-Step: A Bilayer Around Your Protein
Here’s a quick overview of how to use the Molecular Box Builder to surround a protein with lipids:
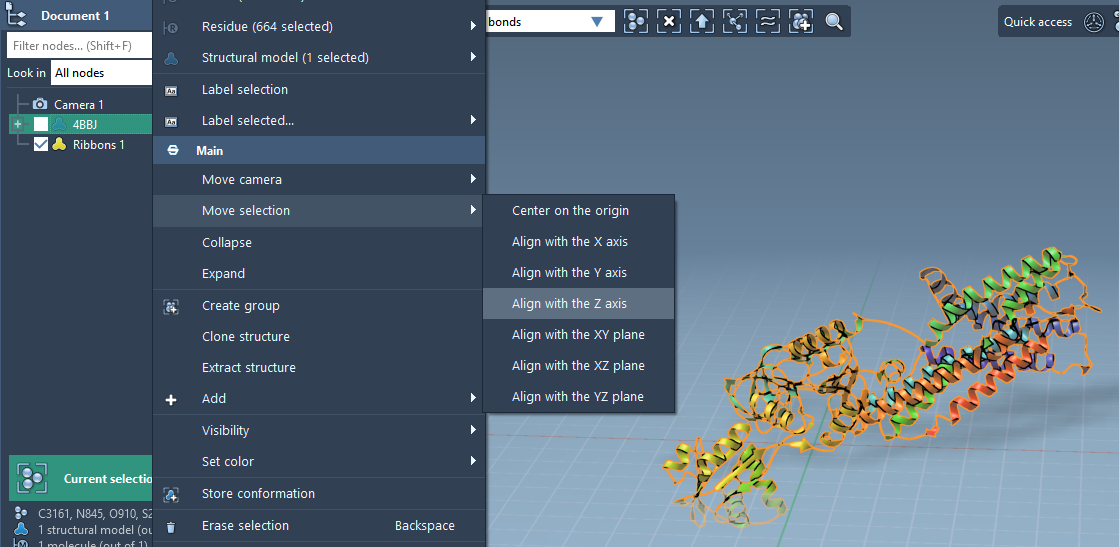
1. Align the Protein
- In the Document view, right-click your protein.
- Select Move selection > Align with Z axis.
- Then click Move selection > Center on the origin.
This ensures that the membrane will form in the XY plane, which matches the typical orientation used in simulations.
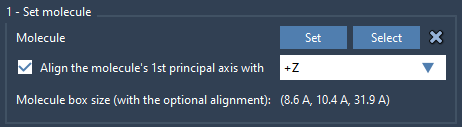
2. Import and Align the Lipid
- Import a lipid molecule of your choice into SAMSON (e.g., POPC, DPPC, etc.).
- Select it in the Viewport or Document view.
- In the app, click Set to mark it as the insertion molecule.
- Choose the +Z orientation to align its principal axis.
3. Define the Box
- Create a bounding box centered on the protein.
- Adjust the dimensions in X and Y to span the desired area of the membrane.
- For a monolayer, keep Z small. For a bilayer, you’ll repeat this step later with a shift.
- Activate Consider existing molecules in the box so the lipids avoid overlapping with the protein.
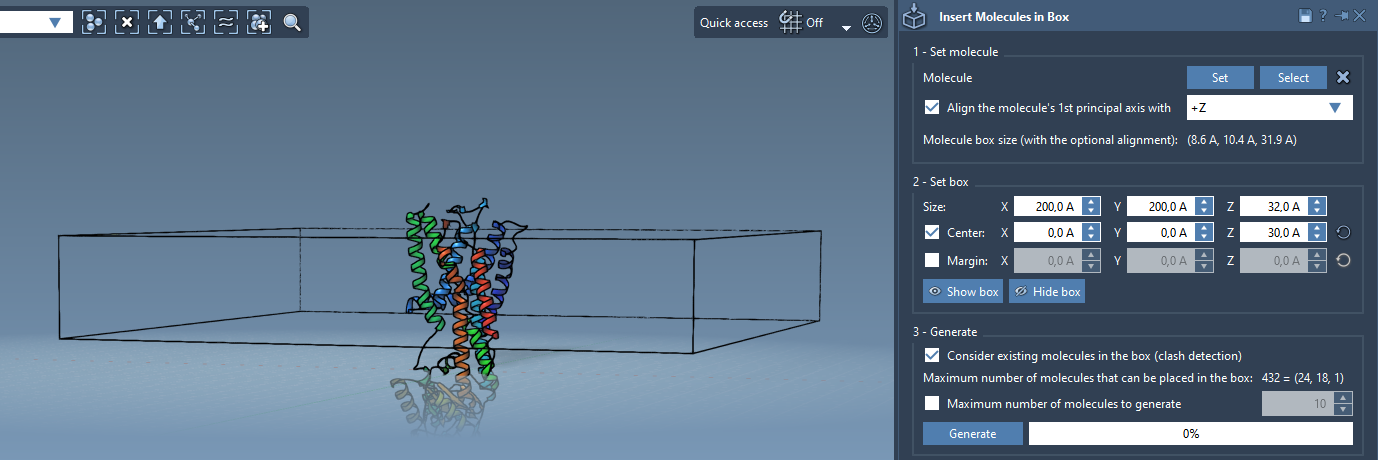
4. Generate the First Layer
- Click Generate to place the first lipid layer on the +Z side.
Already, you have a biologically relevant lipid leaflet.

5. Build the Bilayer
- Shift the box in the Z-direction (e.g., down by 3-4 nm depending on system size).
- Change the lipid orientation to -Z.
- Click Generate again.
Now your system contains a full lipid bilayer around your protein – no scripting or manual alignment required.

What’s Next?
Once built, you can proceed with adding water molecules, ions, and running membrane-protein molecular dynamics simulations. For example, the GROMACS Wizard in SAMSON helps you set up and launch those simulations.
To learn more, visit the full documentation for the Molecular Box Builder: https://documentation.samson-connect.net/tutorials/molecular-box-builder/molecular-box-builder/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





