No Admin Rights? No Problem: Installing SAMSON Without Admin Privileges
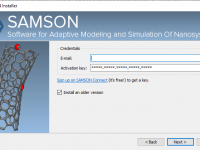
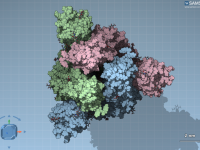
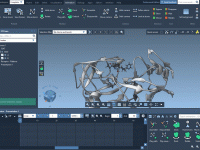
For molecular modelers working in institutional settings—especially students and researchers without administrative privileges on their workstations—software installation can be a persistent challenge. Many powerful scientific tools require admin rights, which slows down onboarding and introduces a frustrating dependency on IT…